Abstract
Gene expression depends on binding of transcriptional regulators to gene promoters, a process controlled by signalling pathways. The transcriptional repressor B-cell lymphoma (BCL)-6 downregulates genes involved in cell-cycle progression and becomes inactivated following phosphorylation by the Rac1 GTPase-activated protein kinase PAK1. Interestingly, the DNA motifs recognized by BCL-6 and signal transducers and activators of transcription 5 (STAT5) are similar. Because STAT5 stimulation in epithelial cells can also be triggered by Rac1 signalling, we asked whether both factors have opposing roles in transcriptional regulation and whether Rac1 signalling may coordinate a transcription factor switch. We used chromatin immunoprecipitation to show that active Rac1 promotes release of the repressor BCL-6 while increasing binding of STAT5A to a BCL-6-regulated reporter gene. We further show in colorectal cell lines that the endogenous activation status of the Rac1/PAK1 pathway correlated with the phosphorylation status of BCL-6 and STAT5A. Three cellular genes (cyclin D2, p15INK4B, small ubiquitin-like modifier 1) were identified to be inversely regulated by BCL-6 and STAT5A and responded to Rac1 signalling with increased expression and corresponding changes in promoter occupancy. Together, our data show that Rac1 signalling controls a group of target genes that are repressed by BCL-6 and activated by STAT5A, providing novel insights into the modulation of gene transcription by GTPase signalling.
INTRODUCTION
A crucial process in gene expression is the initiation of gene transcription. Before ribonucleic acid (RNA) polymerase II can transcribe the coding information of a given gene into RNA, it generally needs to be recruited to the respective gene promoter by specific transcription factors. These factors recognize conserved short DNA sequence motifs in the promoter but usually only bind to them following transcription factor activation and chromatin remodelling. Consequently, transcriptional regulation is frequently preceded by cellular signalling events. For example, activation of growth factor receptors at the plasma membrane stimulates the Ras/Raf/extracellular signal-regulated kinase (ERK) pathway, and activated ERK translocates into the nucleus where it phosphorylates transcription factors such as Elk-1 and Myc, enabling them to bind and activate target gene promoters (1). A different strategy is used by activated cytokine receptors, which stimulate tyrosine phosphorylation of the signal transducers and activators of transcription (STAT) family of transcription factors at the plasma membrane and these activated factors then translocate into the nucleus to activate their target genes (2).
Another signalling molecule activated downstream of membrane receptors is the small guanosine triphosphate (GTPase) Rac1, initially discovered for its ability to stimulate the polymerization of actin filaments and cell migration (3). In addition, Rac1 has distinct roles in the regulation of gene transcription (4). For instance, the stimulation of c-Jun N-terminal kinase by Rac signalling leads to the phosphorylation and subsequent activation of the transcription factors c-jun, activating transcription factor (ATF), ETS-like transcription factor (ELK) or activator protein 1 (AP1). A further transcription factor stimulated by Rac1 signalling is Nuclear factor kappa-light-chain-gene-enhancer of activated B cells (NF-κB) and involves the phosphorylation and proteolytic degradation of the cytoplasmic inhibitor proteins IκBα and NF-κB2/p100 (5,6).
Some STAT factors were also reported to be regulated by Rac1. They form a family of seven transcription factors, are found in the cytoplasm under basal conditions and enter the nucleus following their activation by tyrosine phosphorylation (2). STAT3 binds directly to active Rac1, possibly targeting STAT3 to tyrosine kinase signalling complexes (7). In addition, Rac1 and a GTPase-activating protein, MgcRacGAP, bind directly to phosphorylated STAT3 and STAT5A, promoting their nuclear translocation and activity (8,9).
Previously, we reported a novel link between Rac1 signalling and transcriptional regulation. Rac1 activation leads to p21-activated kinase (PAK1)-mediated phosphorylation of the transcriptional repressor B-cell lymphoma (BCL)-6 in colorectal tumour cells and inactivates its repressor function (10). BCL-6 was initially identified as a repressor gene translocated in B cell non-Hodgkin’s lymphomas (11–13). Later, BCL-6 expression has also been detected in non-haematopoietic tissues, including skeletal muscle (14), uroepithelial cells (15,16), olfactory sensory neurons (17), skin (18), epithelial cells of the mammary gland (19) and HeLa cells (20). BCL-6 contains carboxy-terminal zinc finger modules that bind DNA in a sequence-specific manner (21,22). The genes repressed by BCL-6 are best studied in germinal centre B cells and involved in lymphocyte activation and terminal differentiation, including cell-cycle regulation (12,23–25).
Interestingly, the DNA motifs recognized by BCL-6 are highly homologous to the core binding sequence TTCNNNGAA of STAT factors STAT5 (2,26). This raised the hypothesis that both factors may have opposing roles in the transcriptional regulation of some target genes. Here, we used chromatin immunoprecipitation (ChIP) to show that active Rac1 promotes release of the repressor BCL-6 from promoters together with increased binding of STAT5A. We also identify three endogenous target genes involved in cell-cycle control that were inversely regulated by BCL-6 and STAT5A and responded to Rac1 signalling with a transcription factor switch.
MATERIALS AND METHODS
Cell culture and transfection
DLD-1 and SW480 colorectal cells were maintained in Dulbecco’s minimal essential medium and HT29 cells were kept in Roswell Park Memorial Institute medium (RPMI) medium, both supplemented with 10% (v/v) foetal bovine serum (all from Invitrogen) and regularly checked for absence of mycoplasm infection. Cells were transfected as previously described (10), using a 1:2 proportion (µg/μl) of DNA:LipofectAMINE 2000 (Invitrogen) and total amounts of transfected plasmid DNA of 12 µg per 100-mm dish for ChIP assays and 2 µg per 35-mm dish for cell fractionation, reporter assays and transcript expression analysis in case of DLD-1 and SW480 cells, but twice the amount for HT29 cells. Optimal transfection efficiencies were 60–80% in DLD-1 or SW480 cells and 40–60% in HT29, as judged microscopically by expression of green fluorescent protein (GFP)-tagged vector and cells analysed 16–20 h later.
For RNA interference experiments, cells were transfected at 20–40% confluence in 35-mm dishes with either 200 pmol (DLD-1) or 400 pmol (HT29) of the indicated siRNAs using LipofectAMINE 2000 and analysed 48 h later. The siRNA oligos against BCL-6 (sc29791), STAT5A (sc-29495) and PAK1 (sc-29700) were from Santa Cruz Biotechnology and a scramble control oligo (5′-AGG UAG UGU AAU CGC CUU GTT) from Eurofins MWG Operon.
DNA plasmids and constructs
The following published constructs were received as gifts: PAK1-wt, kinase-dead dominant negative PAK1-K299R and constitutively active PAK1-T423E mutants from J. Chernoff (Philadelphia), and the 5xBCL-6-vector (22) from V.J. Bardwell (Minnesota). Myc-Rac1, GFP-Rac1 and pEGFP-BCL-6 constructs were previously described (6,10,27). STrEP-tagged BCL-6 was generated by subcloning a BamHI/Xho I fragment from pcDNA3-BCL-6 into vector pEXPR-IBA105 (IBA, Göttingen, Germany). pEGFP-STAT5A was generated by polymerase chain reaction (PCR) amplification of the STAT5A cDNA from pMX-STAT5A (gift from B. Groner, Frankfurt) using a forward primer (5′-ATG GCG GGC TGG ATT CAG G) and a reverse primer (5′-ATC TCA GGA CAG GGA GCT TCT) and subcloned into pEGFPc2 using EcoRI restriction sites. All constructs were confirmed by automated DNA sequencing.
Analysis of transcript expression by quantitative reverse transcriptase-PCR
Total RNA was extracted from cell lysates with the NucleoSpin RNA kit (Macherey–Nagel) and 1 µg reverse transcribed using random primers (Invitrogen) and Ready-to-Go You-Prime First-Strand Beads (GE Healthcare). CCND2, CDKN2B and small ubiquitin-like modifier 1 (SUMO1) transcript levels were determined by quantitative PCR (qPCR) on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using the primers and PCR conditions summarized in Supplementary Table S1. Each cDNA sample was diluted 5-fold to guarantee accurate pipetting and 5 µl added to each real-time reaction together with 200 nM primers and SYBR Green Master Mix (Applied Biosystems). Data were analyzed with the 7000 SDS 1.1 RQ Software (ΔΔCT method, Applied Biosystems) (28) using mock transfections as reference samples. For comparison of gene expression between cell lines a pool of cDNAs mixed at equal parts from the three cell lines was used as reference.
Semi-quantitative reverse transcriptase-PCR was used to estimate siRNA-mediated knockdown of BCL-6 and STAT5A expression. RNA polymerase II (as earlier) was amplified as a control gene and two serial dilutions of scramble siRNA sample served to assure semi-qPCR conditions and estimate knockdown efficiency.
PCR array analysis
The Human Cell-Cycle PCR array (PAHS-020, SABiosciences/Qiagen) was used according to manufacturer’s instructions. An RNA pool from three independent siRNA experiments performed in DLD-1 or HT29 cells was reverse transcribed, then added to a SYBR Green/Rox qPCR Master Mix (PA-012, Qiagen), distributed into the 96-well array plate and measured by qPCR as described previously. The quantitative analysis was done on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) with the following cycling conditions: 10 min at 95°C and 40 cycles at 95°C for 15 s and 60°C for 60 s. Two PCR arrays were used for each experimental condition. Data analysis was performed using the Excel-based tool provided by the manufacturer.
Identification of putative BCL-6/STAT5 binding sites
A 2500 bp of the genomic sequence immediately upstream the annotated transcription initiation sites for CCND2, CDKN2B and SUMO1 genes were used to search in silico for putative binding sites recognized simultaneously by BCL-6 and STAT5A. Several algorithms were employed (http://www.gene-regulation.com/; http://www.biobase-international.com/; http://www.genomatix.de) using the score values obtained for the previously described BCL-6/STAT5 site in CCND2 (29) as a reference for parameter adjustment and best putative site selection.
Chromatin immunoprecipitation
When indicated, DLD-1, SW480 or HT29 cells were transfected with expression vectors and assayed 16 h later. ChIP was performed as previously described (29). Briefly, approximately 10 x 106 cells per ChIP were cross-linked with 1% formaldehyde, lysed and sonicated to produce chromatin fragment between 200 and 500 bp (40% power on a Sonics Vibra Cell sonicator). Cleared samples were incubated overnight at 4°C with either anti-BCL-6 N3 (Santa Cruz Biotechnologies), anti-STAT5A (Invitrogen) or control anti-rabbit immunoglobulin G (DakoCytomation; P 0448) antibodies, preserving 1/10 lysate volume as input control. Protein G-conjugated magnetic beads (Invitrogen) were then added for 1 h at 4°C. Beads were thoroughly washed and co-precipitated DNA was purified with the QIAquick PCR purification kit (Qiagen) after cross-link reversion. The selected putative STAT5A/BCL-6 binding regions were amplified from ChIP samples, with the primers and conditions described in Supplementary Table S1. As a specificity control, a genomic fragment between intron 8 and intron 10 of the MutY homolog (MUTYH) gene (Accession number NG_008189) was amplified. Products were separated on 2% agarose gels containing ethidium bromide. Two serial dilutions of the “input DNA” control were co-amplified to guarantee semi-qPCR conditions and allow product quantity extrapolation from band intensities analysed on digital images using ImageJ software (National Institutes of Health), which were then normalized to the control sample.
SDS-PAGE, Western blotting and Rac pull-down assays
Samples were prepared and detected as described (10). The antibodies used for Western blots were rabbit polyclonal anti-c-Myc A14, anti-Histone H2B (sc-10808) and rabbit anti-BCL-6 (clone N3, sc858) from Santa Cruz Biotechnology, monoclonal anti-GFP (#11814460001) from Roche, monoclonal anti-Rac1 (clone 23A8) from Upstate Biotechnologies (#05-38), anti-PAK1 ab40795 from Abcam, polyclonals anti-phospho-PAK1 (Ser199/204)/PAK2 (Ser192/197) (#2605) and anti-phospho-STAT5A (Tyr694) (#9351) from Cell Signalling Technology, monoclonal anti-STAT5A from Invitrogen (#13-3600) and monoclonal anti-α-tubulin clone B-5-1-2 (as loading control) from Sigma-Aldrich (T6074). The Rac pull down assay was as described (6). For densitometric analysis, films from at least three independent experiments were digitalized and analysed using ImageJ software (National Institutes of Health).
Cell fractionation
Proteins were separated into a soluble pool not retained in the nucleus and into a chromatin-bound insoluble pool according to previously described procedures (10,30). Briefly, cells were washed in cold PBS, scraped off and lysed on ice in fractionation buffer [50 mM Tris-HCl, pH 7.9, 0.1% (v/v) NP40, 1.5 mM MgCl2, 10 mM KCl and a protease inhibitor cocktail (Sigma)]. The soluble fraction was collected by centrifugation and adding the supernatant to 5× Laemmli SDS sample buffer. The pellet containing the insoluble fraction in nuclei was washed once in fractionation buffer and then resuspended in 1× Laemmli sample buffer. Equal volumes of both fractions were analysed side by side on Western blots. Results were confirmed in at least three independent experiments.
Confocal immunofluorescence microscopy
Experiments were performed as described (10) and images recorded with a Leica TCS-SPE confocal microscope.
Luciferase Reporter Assay
The use of the pGL3-5xBCL-6 reporter vector in DLD-1 cells was previously described (10). Briefly, cells were co-transfected with pRL-TK luciferase reporter (internal control; Promega), pGL3-5xBCL-6 or pGL3 control reporters, and the indicated expression constructs. After 16–20 h cells were lysed and assayed with the Dual Luciferase Reporter Assay (Promega) and measured in an Anthos Lucy-2 Luminometer. Lysates were assayed in duplicates and additional aliquots analysed by Western Blot to document protein expression levels. Normalized luciferase values were plotted as fold-increase over the value of control treatments and correspond to at least three independent transfection assays.
Statistical analysis
Statistical significance of the differences between treated and control samples was analyzed using two-tailed Student’s t-tests and indicated in the figures by an asterisk (*) whenever p < 0.05.
RESULTS
Rac1 signalling promotes transcription by repressing BCL-6 and stimulating STAT5
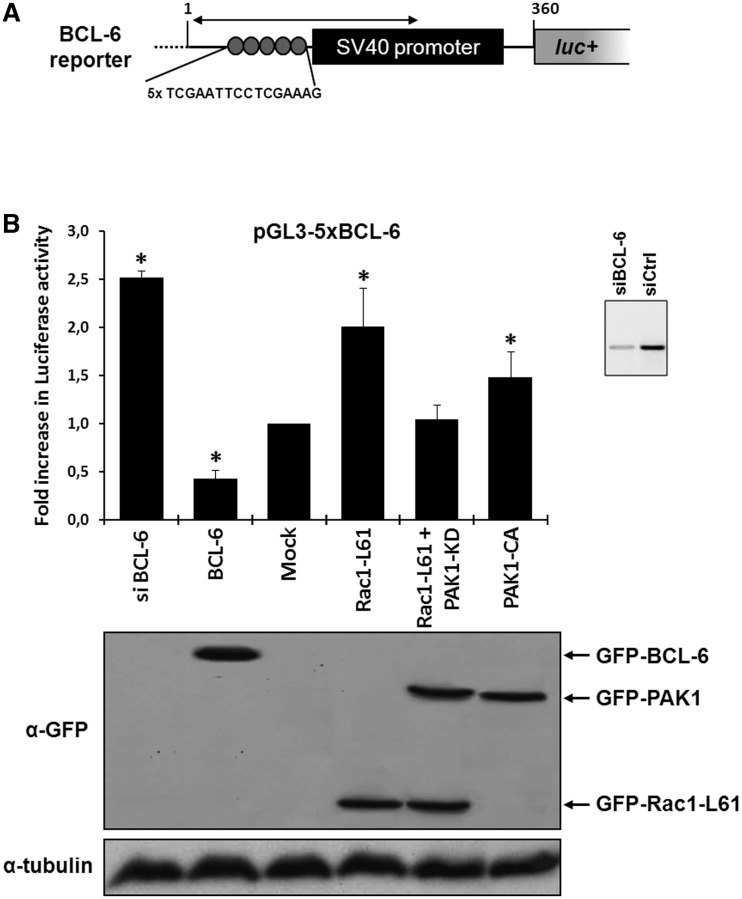
Recently, we used a BCL-6 reporter gene construct (Figure 1A) in which five repeats of a BCL-6 recognition motif control luciferase expression and found that Rac1 signalling acts as an upstream regulator of BCL-6 in colorectal DLD-1 cells (10). When this reporter gene was transfected into DLD-1 cells together with GFP-tagged BCL-6, a further repression was observed, whereas depletion of endogenous BCL-6 expression by RNA interference led to transcriptional activation (Figure 1B). In the course of these studies, we noticed that the expression of active Rac1-Q61L had a stronger stimulatory effect on reporter gene transcription than a constitutively active PAK1-T423E mutant (Figure 1B), although PAK1 is activated downstream of Rac1 and was shown to phosphorylate BCL-6 (10). We thus reasoned that Rac1 may activate additional PAK1-independent pathways that affect reporter gene activation. One candidate pathway was activation of STAT5 because STAT5 was reported to recognize BCL-6 binding motifs in some cellular genes, including cyclin D2 or prolactin (25,29,31–33), and because it formed a complex with active Rac1 promoting STAT5 nuclear import and transcriptional activation (9).
Figure 1.
Rac1 signalling promotes transcription by repressing BCL-6 and stimulating STAT5. (A) Schematic representation of the transcriptional luciferase reporter vector under the control of five consensus BCL-6 binding motifs. (B) DLD-1 cells were co-transfected with the reporter vector and the indicated expression vectors or siRNAs. Cells were lysed 24 h later and luciferase activity was measured and graphically displayed, *P < 0.05. The Western blot below the graph shows the levels of transfected GFP-tagged BCL-6, Rac1-L61, PAK1 kinase dead (kd) or PAK1 constitutively active (ca) mutants. Detection of endogenous α-tubulin served as loading control. The small insert beside the graph shows a Western blot of endogenous BCL-6 to document the efficiency of its siRNA-mediated depletion.
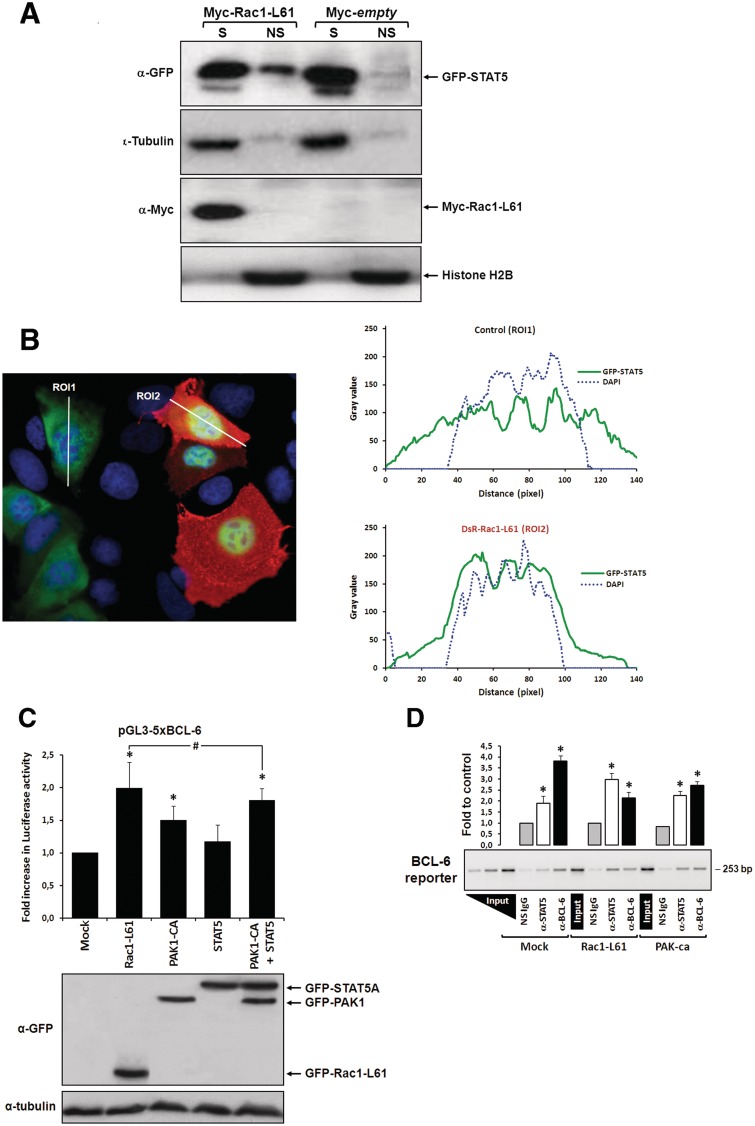
To test whether active Rac1 could promote nuclear translocation of STAT5 in DLD-1 cells, we first applied a cell fractionation protocol, which separates transcription factors into a soluble pool that is extracted from the nucleus and a chromatin-bound pool that remains insoluble (10). Under control conditions, STAT5 was detected in the soluble fraction (Figure 2A), whereas in cells co-expressing an active Rac1-Q61L mutant a notable transition of STAT5 into the chromatin-bound insoluble fraction was observed (Figure 2A). Second, we visualized the effect of active Rac1 on STAT5 by fluorescence microscopy in cells co-transfected with DsRed-Rac1-Q61L and GFP-STAT5A. As shown in Figure 2B, a clear transition of STAT5 into the nucleus was observed.
Figure 2.
Rac1 signalling switches promoter occupancy from BCL-6 to STAT5. (A) DLD-1 cells were transfected with Myc-Rac1-Q61L or control empty vector and 24 h later analyzed by Western blot for the subcellular distribution of STAT5 between a soluble (S) and a chromatin-bound non-soluble (NS) fraction (detection of α-tubulin and histone 2B served as controls). Note that active Rac1 promotes retention of STAT5 in the non-soluble chromatin fraction. (B) Subcellular localization of STAT5 determined by confocal fluorescence microscopy in DLD-1 cells co-transfected with DsRed-Rac1-Q61L and GFP-STAT5A. The overlay image of the DAPI, GFP and DsRed channels is shown. A microscopic field was chosen that contained side by side untransfected cells (blue nuclei), cells that transfected only with GFP-STAT5 (green cells) and cells that co-transfected with both GFP-STAT5 and DsRed-Rac1-Q61L (red cells). Note the nuclear STAT5 signal in Rac1-expressing red cells. In addition, two plots are given showing the DAPI and GFP signal intensities measured along the indicated regions of interest (ROI, white lines). The signal intensity of GFP did not increase across the nuclear DAPI region when cells expressed only GFP-STAT5 (green cells, ROI 1), whereas nuclear GFP signal clearly increased when cells co-expressed active Rac1 (red cells, ROI 2), confirming nuclear translocation of GFP-STAT5. (C) DLD-1 cells were co-transfected with the reporter and the indicated expression vectors, as described for Figure 1B. Note that STAT5 activates the BCL-6 luciferase reporter and, when combined with PAK1, reaches the stimulation levels normally induced by active Rac1, *P < 0.05 and #P > 0.05. (D) Chromatin immunoprecipitation (ChIP) of the reporter vector with anti-(α)-BCL-6, α-STAT5 or a non-specific antibody (NS immunoglobulin G) from lysates of DLD-1 cells transfected with the indicated expression vectors. A representative semi-qPCR of the precipitated promoter fragment quantities with a graphical representation of the respective band intensities quantified by densitometry from digital images obtained in 3 independent transfection experiments, *P < 0.05, is shown. Two serial dilutions of input DNA were co-amplified to guarantee semi-qPCR conditions and allow product quantity extrapolation from band intensities.
To confirm whether STAT5 was able to activate the BCL-6 reporter gene under these conditions, both constructs were co-transfected into DLD-1 cells and increased luciferase transcription was measured (Figure 2C). We then co-expressed STAT5 and constitutively active PAK1 to test whether their combined transcriptional activation would mimic that induced by Rac1-Q61L. As shown in Figure 2C, simultaneous stimulation of PAK1 and STAT5 could indeed account for the complete stimulatory effect induced by Rac1 signalling.
Rac1 signalling switches promoter occupancy from BCL-6 to STAT5
These results suggested that Rac1 signalling activates two independent pathways of transcriptional regulation that target the same reporter gene. To obtain further support for this conclusion, we determined the occupancy of the reporter gene promoter by either BCL-6 or STAT5 under the various experimental conditions. For this, DLD-1 cells were co-transfected with the BCL-6 reporter gene and either control vector or active Rac1-Q61L or active PAK1-T423E and the presence of either transcription factor at the reporter gene promoter was analysed by ChIP. As shown in Figure 2D, BCL-6 was the predominantly bound factor in control cells, however, upon expression of active Rac1 BCL-6 binding was reduced and STAT5 became the predominantly bound factor at the promoter. To exclude that the observed changes in promoter occupancy were the result of epitope masking (due to an interaction of BCL-6 and STAT5 at the promoter that could interfere with recognition by their specific ChIP antibodies), co-precipitation studies were carried out (Supplementary Figure S1). No evidence was found for complex formation between the two transcription factors, indicating that changes in promoter occupancy reflected changes in bound proteins. In case of active PAK1, BCL-6 was also partially reduced, and this is in agreement with our previous data that PAK1 phosphorylates BCL-6 and promotes its release from chromatin and loss of repressor activity. However, in contrast to active Rac1, PAK1 was unable to invert the promoter occupancy from BCL-6 to STAT5. Together, these data support the conclusion that Rac1 signalling activates two independent pathways to promote a switch in promoter occupancy from BCL-6 to STAT5.
Correlation of Rac1 signalling and activation of BCL-6 or STAT5 in different cell lines
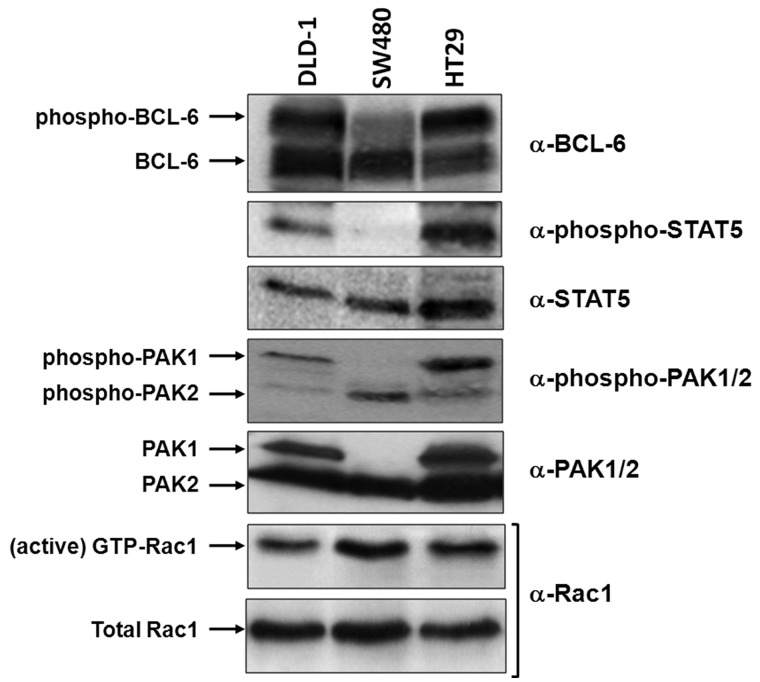
To understand the physiological relevance of the observed transcriptional switching at the reporter gene, we first characterized the endogenous activity levels of Rac1, PAK1, STAT5 and BCL-6 in three different colorectal cell lines using Western blot analysis. As shown in Figure 3, SW480 cells revealed the strongest endogenous Rac1 activation level, followed by HT29 and DLD-1 cells. Curiously, SW480 cell lost PAK1 expression, whereas in HT29 and DLD-1 cells, active Rac1 was proportional to active PAK1, as well as to the levels of phospho-BCL-6 and phospho-STAT5. Interestingly, SW480 cells expressed BCL-6 as well as STAT5 but lacked any significant activation by phosphorylation. This suggested that repression by BCL-6 should be predominant in these cells, indicating their usefulness as a negative control for the transcriptional switch to STAT5 in subsequent experiments.
Figure 3.
Correlation of Rac1 signalling and activation of PAK1, BCL-6 or STAT5 in different cell lines. Equivalent lysate quantities of DLD-1, SW480 and HT29 colorectal cells were separated by gel electrophoresis and analysed by Western blot using the indicated antibodies to compare protein levels. The active Rac1 fraction was obtained by CRIB-pull down assays, as described (6).
Identification of endogenous genes inversely regulated by BCL-6 and STAT5
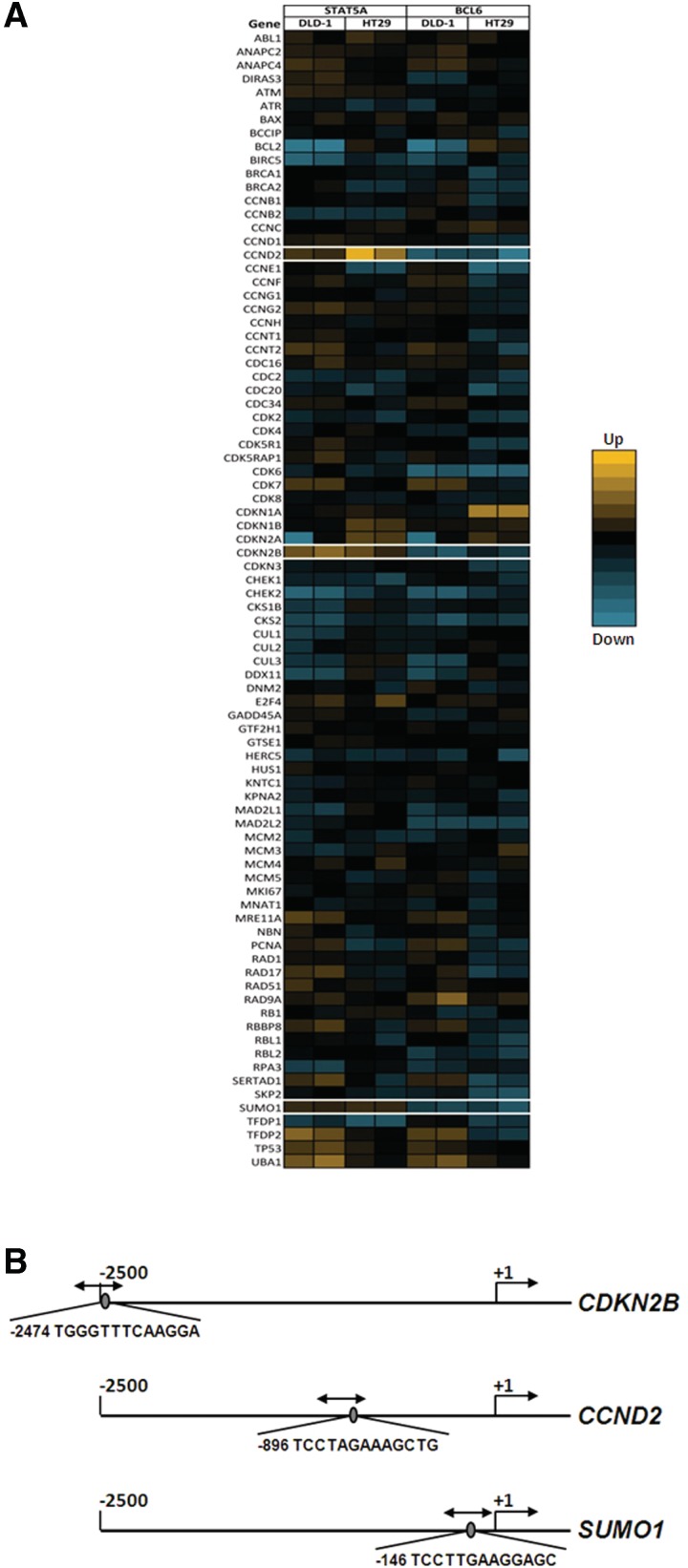
As a next step to identify physiological targets of the observed transcriptional switching, an array of 84 cell-cycle-related genes was tested for opposite effects of BCL-6 and STAT5 on gene expression. For this, the two cell lines that showed endogenous BCL-6 and STAT5 activation, DLD-1 and HT29, were independently transfected with small interfering RNAs targeting either BCL-6 or STAT5 (Supplementary Figure S2). qPCR analysis of the resulting gene expression levels identified three genes that were affected in opposite sense by the downregulation of either BCL-6 or STAT5, namely cyclin D2 (CCND2), cyclin-dependent kinase inhibitor p15INK4B (CDKN2B), and SUMO1 (Figure 4A).
Figure 4.
Identification of endogenous genes inversely regulated by BCL-6 and STAT5. (A) DLD-1 and HT29 cells were transfected with either BCL-6 or STAT5-specific siRNA oligonucleotides and lysed following 48 h for RNA extraction. (Supplementary Figure S2). A heat map display of the gene expression analysis of a cell-cycle PCR array probed with RNA samples obtained from BCL-6 or STAT5-depleted DLD-1 and HT29 cells, is shown. Of the 84 genes on the array, 3 were identified to be regulated by BCL-6 and STAT5 in opposite sense (white boxes). (B) Schematic representation of the promoter regions of the three endogenous genes inversely regulated by BCL-6 and STAT5 showing the selected best putative motifs, with equivalent predicted binding scores for both factors.
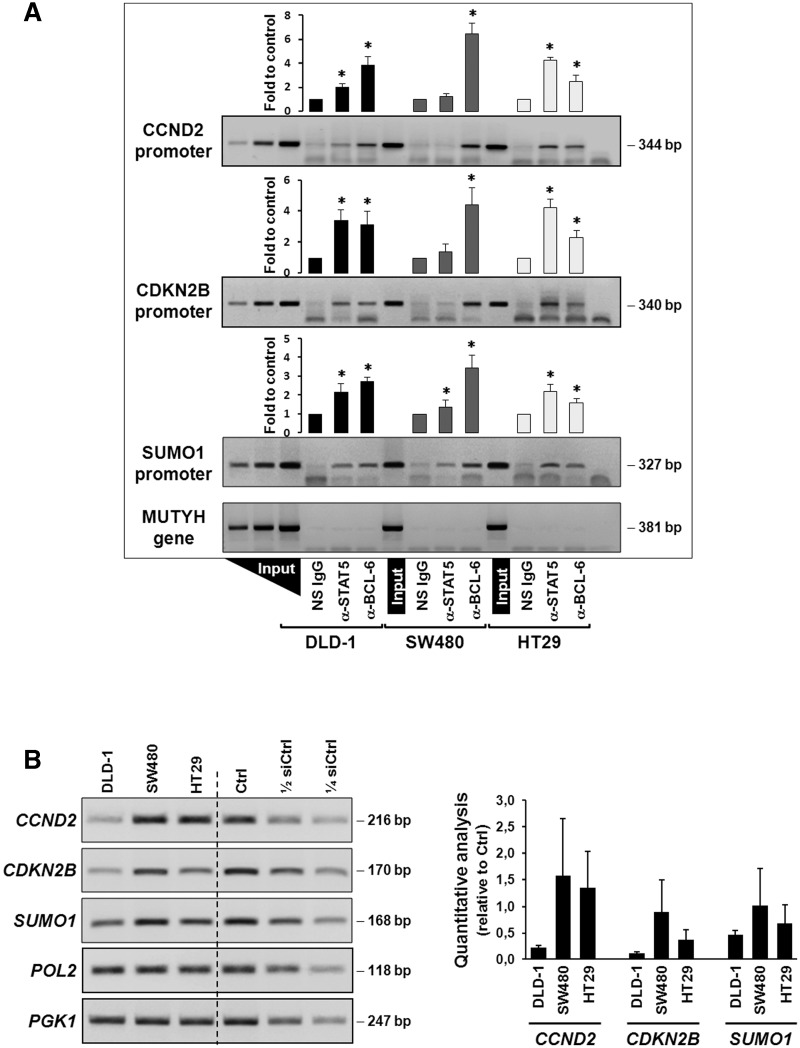
To determine the respective promoter occupancies in these three genes, overlapping binding motifs for BCL-6 and STAT5 were identified using a Transfac® database-based algorithm (see Methods section and Figure 4B) and used to design ChIP experiments in the three aforementioned cell lines. It was found that both factors were bound to the 3 identified promoters regions, albeit to different extent; however, not to a control genomic fragment. In SW480 cells (that express no PAK1 and have little phospho-BCL-6; Figure 3), the predominant factor that was bound to all 3 promoters was BCL-6, whereas STAT5 was close to background levels (Figure 5A, grey bars). In contrast, HT29 cells contained more STAT5 bound to these promoters than BCL-6 (Figure 5A, white bars), in agreement with their higher endogenous levels of active PAK1, phospho-BCL-6 and phospho-STAT5 (cf. Figure 3). In DLD-1 cells (Figure 5A, black bars), comparable promoter binding levels were detected for both factors (except for the cyclin D2 promoter that had more BCL-6 bound). Again, this is in good agreement with the observation described in Figure 3 that endogenous levels of active PAK1, phospho-BCL-6 and phospho-STAT5 in DLD-1 were lower than in HT29 but higher than in SW480 cells (Figure 3).
Figure 5.
Regulation of expression of the CCND2, CDKN2B and SUMO1 genes. (A) Promoter occupancies with BCL-6 and STAT5 at the CCND2, CDKN2B and SUMO1 gene promoters was determined by ChIP with the indicated antibodies using lysates of the three indicated cell lines (see legend to Figure 2D for details). A representative semi-qPCR of the precipitated promoter fragments and a graphical representation of the respective band intensities, *P < 0.05, are shown. A control genomic fragment from the MUTYH gene was amplified to confirm the specificity of the precipitated target gene promoters. Note that BCL-6 binds predominantly in the PAK1-lacking SW480 cells and whereas a switch to STAT5 occurs in HT29 cells with active Rac1/PAK1 signalling. (B) Gene expression data corresponding to the ChIP analysis of the three genes in the three cell lines. Left panel shows representative semi-quantitative reverse transcriptase-PCRs (RT-PCRs), whereas graph at the right shows the result of qPCR analysis of cDNAs collected from the three cell lines at three different splitting times. Genes encoding RNA polymerase II (pol 2) and the glycolytic enzyme phosphoglycerate kinase (PGK1) were amplified as control housekeeping genes and a pool of cDNAs mixed at equal parts from the three cell lines was used as reference for qPCR. Serial dilutions served to assure semi-quantitative conditions in the conventional RT-PCR reactions.
Next, these data on the promoter occupancies of the CCND2, CDKN2B and SUMO1 genes were matched to the corresponding gene expression levels, validated by qPCR using independently designed PCR primers (Figure 5B). HT29 cells that had less BCL-6 repressor bound than DLD-1 cells also revealed higher expression levels for all three genes. Surprisingly, SW480 cells also expressed all three genes considerably, although BCL-6 was predominantly bound in these cells, indicating they use different mechanisms to activate these cell-cycle regulating genes.
Rac1 signalling controls reciprocal roles of BCL-6 and STAT5 in target gene expression
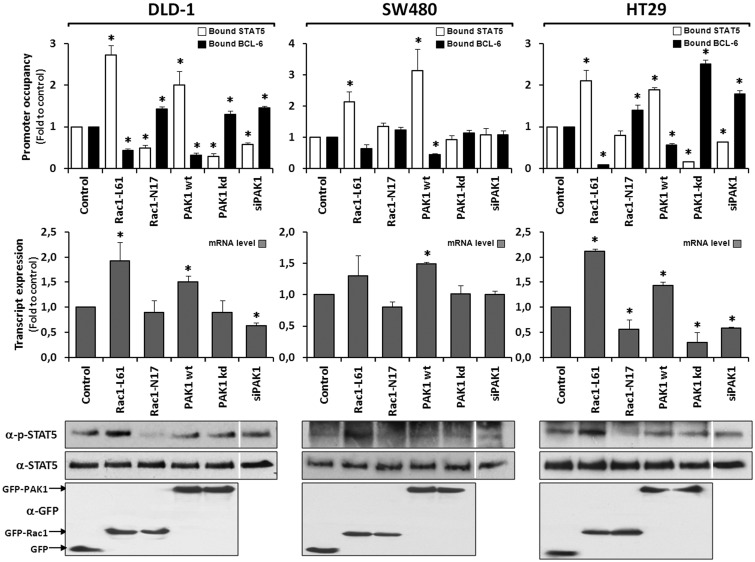
As final evidence that the transcription factor switch is physiologically meaningful, the promoter occupancies at the 3 genes were determined and compared with changes in their respective expression levels following activation or inhibition of Rac1 signalling in the 3 different cell lines. For this, cells were transfected with vectors encoding either PAK1, or dominant negative or active Rac1, or with siRNA oligonucleotides directed against endogenous PAK1 (depletion documented in Figure S2B). The 3 genes revealed equivalent results, which are represented in Figure 6 for the SUMO1 gene by displaying the levels of promoter-bound BCL-6 or STAT5 alongside the respective target gene transcript levels. Comparable data for the CCND2 and CDKN2B genes are shown in Figures S3 and S4, respectively. When PAK1 was transfected into SW480 cells, which lack endogenous PAK1, a loss of BCL-6 from the promoter of all three genes was induced, which slightly increased their expression levels. By contrast, the depletion of endogenous PAK1 had no effect on promoter occupancy or gene expression (Figure 6 middle panel), a result in agreement with the fact that no endogenous PAK1 is expressed in SW480 cells. When SW480 cells were transfected with active Rac1, a small increase in STAT5 phosphorylation and binding to the promoter was observed; however, the overall effect on gene expression was negligible because the lack of PAK1 compromised BCL-6 removal from the promoter. These data confirm our previous assumption that SW480 cells represent a negative control and cannot respond to Rac1 signalling with the transcriptional switch between BCL-6 and STAT5.
Figure 6.
Rac1 signalling controls target gene expression by inverting promoter occupancy with either BCL-6 or STAT5. The representative analysis of the SUMO1 gene is shown in the indicated three colorectal cell lines following their transfection with constructs that either activate or inhibit Rac1 signalling. Top panels show the graphical display of promoter occupancy by ChIP using either anti-BCL-6 (black columns) or STAT5 (white columns) and middle panels the respective gene expression levels (grey columns) (see legend to Figure 5 for further details), *P < 0.05. Bottom panels show Western blot analysis of the cell lysates demonstrating the expression levels of the transfected GFP, GFP-Rac1 or GFP-PAK1 constructs as well as the resulting phosphorylation status of endogenous STAT5. Note that in SW480, which lack endogenous PAK1, depletion of endogenous PAK1 by siRNAs transfection (documented in Fig. S2B) or expression of dominant negative PAK1 has no effect on promoter-bound BCL-6, whereas re-expression of PAK1 leads to loss of BCL-6 from the SUMO1 promoter and an increase in gene expression. In the other two cell lines, inhibition of Rac1 or PAK1 are clearly correlated with more BCL-6 bound and less gene expression while activation of Rac1 or PAK1 promoted STAT5 binding to the promoters and increased transcription.
In contrast, HT29 and DLD-1 cells both switched BCL-6 and STAT5 at the three gene promoters upon transfection with active Rac1, accompanied by a clear increase in STAT5 phosphorylation and in gene expression. Upon transfection of these cells with PAK1, BCL-6 was lost from the three gene promoters and expression increased slightly; however, no significant increase in STAT5 phosphorylation occurred. In the presence of PAK1-specific siRNAs (as well as a dominant negative PAK1 mutant), BCL-6 promoter occupancy increased and expression of the three genes was inhibited.
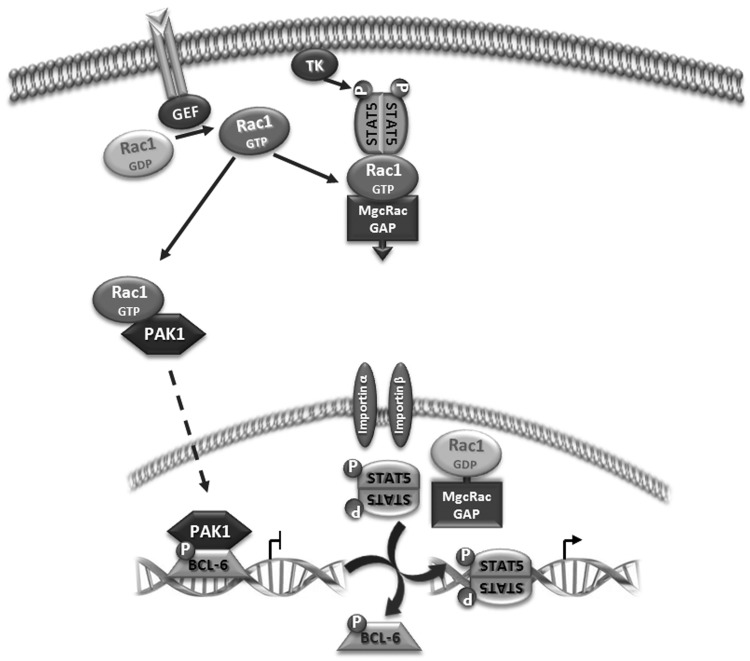
Altogether, these data provide evidence for the model proposed in Figure 7, showing that Rac1 signalling has a dual effect on transcriptional regulation of the CCND2, CDKN2B and SUMO1 genes. First, Rac1 activates PAK1 which phosphorylates BCL-6 leading to its removal from the target gene promoter and a concomitant increase in gene expression. In parallel, Rac1 activates phosphorylation and nuclear translocation of STAT5, which binds to the same sequence motif in the gene promoter that is recognized by BCL-6 and further increases gene expression.
Figure 7.
Proposed model for the role of Rac1 signalling in the observed transcriptional switch. On receptor activation, exchange factors (GEF) promote GTP binding of Rac1 that stimulates two independent pathways. Active Rac1 binds and activates protein kinase PAK1 that migrates into the nucleus and phosphorylates chromatin-bound BCL-6, leading to its inactivation and loss of promoter occupancy. In parallel, a protein complex is formed between active Rac1, MgcRacGAP and STAT5, promoting phosphorylation by a tyrosine kinase (TK) and translocation into the nucleus. Here, MgcRacGAP stimulates GTP hydrolysis by Rac1 and phospho-STAT5 is released and activates gene transcription following binding to the vacant promoter sites previously repressed by BCL-6.
DISCUSSION
The main finding in this work is that Rac1 signalling activates gene transcription by inducing a switch from repressor BCL-6 to activator STAT5 at the promoter of certain cellular target genes in colorectal cells.
Although BCL-6 is best known as a regulator of B lymphocyte growth and differentiation, it is also expressed in epithelial tissues including skin (18), the mammary gland (19), HeLa cells (20) and colorectal cells (10). Similarly, STATs were described as integral parts of cytokine signalling pathways in haematopoietic cells (2), but meanwhile their role in epithelial cancers has been well documented (34). In particular, aberrant activation of STAT5 was found in prostate (35) and colorectal cancer (36). In these cases, the activation of STAT5 can be mediated by Rac1 signalling, either through the production of reactive oxygen species downstream of G-protein coupled receptor stimulation, leading to activation of the tyrosine kinase JAK (37) and/or through complex formation with MgcGAP promoting nuclear import of phospho-STAT5 (9). Indeed, our study in the colorectal cell lines confirmed that activated Rac1 led to increased phosphorylation of STAT5 (Figure 6) and an increase in chromatin-bound nuclear STAT5 (Figure 2A).
Previous reports have suggested that STAT5 and BCL-6 could bind in a mutually exclusive manner to the same sequence motif in the promoters of certain target genes (25,29,31–33). Our data clearly support these studies and show, side-by-side, that the switch in promoter occupancy between BCL-6 and STAT5 correlates directly with changes in gene expression of either a BCL-6-controlled luciferase reporter vector or of three endogenous gene promoters. More importantly, we show for the first time that this switch is regulated by Rac1 signalling and occurs in colorectal tumour cells. Several pieces of evidence contributed to these data. First, ChIP assays revealed that BCL-6 and STAT5 were bound to the identified gene promoters in the three colorectal cell lines. Second, the endogenous activation status of Rac1, PAK1, and phosphorylated BCL-6 or STAT5 correlated well with promoter occupancies in the cell lines, without detectable changes in the total amount of STAT5 or BCL-6 proteins. Third, experimental activation of Rac1 promoted STAT5 phosphorylation and accumulation in the chromatin-bound nuclear fraction. Fourth, the transcript expression levels of the three endogenous genes mirrored their promoter occupancies and responded to activation or inhibition of upstream Rac1 or PAK1 signalling.
As described earlier, the three colorectal cell lines studied differed in their endogenous activation levels of Rac1 signalling and the resulting inhibition of BCL-6 or stimulation of STAT5. SW480 cells apparently lost PAK1 expression and therefore are unable to phosphorylate BCL-6, except when transfected with ectopic PAK1 (Figure 6). Unexpectedly, these cells still revealed significant expression of the CCND2, CDKN2B and SUMO1 genes, which we identified as inversely regulated target genes for BCL-6 and STAT5. This experimental observation indicates that other mechanisms for transcriptional activation of CCND2, CDKN2B and SUMO1 exist and were used by these cells. Because the control of gene expression involves combinatorial patterns of transcription factor binding, the inhibitory effect of BCL-6 was most likely overcome in SW480 cells by other transcription factors that respond to different signalling inputs. For example, the ability of Myc to induced CCDN2 as well as CDKN2B expression has been reported (38,39) and SW480 cells carry an oncogenic mutation in the KRAS gene (28), a strong activator of several signalling pathways.
In contrast, HT29 and DLD-1 cells shared the same regulatory pattern of BCL-6 inhibition and STAT5 activation, differing only in the extent of BCL-6 inhibition, which was more pronounced in HT29 cells. However, on transfection of active Rac1 or PAK1 mutants, the resulting transcriptional stimulation became almost identical in both cell lines. The same was true for the strong inhibitory effect after depletion of endogenous PAK1 by RNA interference or transfection of a dominant-negative PAK1 mutant, whereas SW480 cells did not respond to either treatment. Together, these data provide substantial evidence that Rac1 signalling promotes a switch at the targeted promoters with a release of BCL-6 and enhanced binding of STAT5 to the same site.
Of the 84 cell-cycle related genes analysed, 3 (3.6%) were clearly identified as inversely regulated by BCL-6 and STAT5. CCND2 encodes cyclin D2, which functions as a regulatory subunit of CDK4 or CDK6 required for cell-cycle G1/S transition. CCND2 overexpression has been reported in colorectal tumours and cell lines (40,41).
The SUMO1 gene encodes a small ubiquitin-like protein that can be covalently attached to proteins as a monomer or a lysine-linked polymer. Unlike ubiquitin, sumoylation is not involved in proteolytic degradation of the attached protein but rather modulates nuclear transport or transcriptional regulation (42).
CDKN2B encodes the cyclin-dependent protein kinase inhibitor protein p15 encoded by the INK4b locus, which can form a complex with CDK4 or CDK6, and prevent their activation by cyclin D. Although CCND2 and SUMO-1 overexpression are consistent with the pro-proliferative role usually associated with increased Rac1 signalling, the role of CDKN2B during colorectal cancer progression remains unclear. Intriguingly, the expression of p15 was also found significantly increased in higher grade prostate carcinomas (43), indicating that alternative mechanisms may exist to inactivate its inhibitor function.
Although the particular functional consequences require further clarification, our findings provide a mechanistic model for how Rac1 signalling promotes switching between transcription factors (Figure 7). Beyond the rapid regulation by Rac1 signalling, the described interplay between STAT5 and BCL-6 is likely also modulated at the long term because STAT5 was found to act as a transcriptional repressor on the BCL-6 gene itself (44). In addition, STAT5 has been described to act as a transcriptional repressor on other genes (45,46). This underlines the requirement for a genome-wide study to understand which genes are activated or repressed by BCL-6 or STAT5 alone, and which genes are regulated reciprocally by the switch between both factors that is described in this manuscript. These differences could reside in the sequence motifs of the corresponding promoters or be mediated by the binding of additional protein factors. Our data are thus a contribution to uncover how Rac1 signalling shapes gene expression and how the deregulation of Rac1 activity that is observed for example in cancer (47) promotes cell proliferation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
Fundação para a Ciência e Tecnologia, Portugal [PPCDT/SAU-OBS/57660/2004] (to P.J.), [PTDC/SAU-GMG/119586/2010] (to P.M.), [PEst-OE/BIA/UI4046/2011] (to the BioFig research unit), fellowship [BD 29789/2006] (to P.B.) and contract [Ciência2007] (to P.M.). EMBO fellowship [ASTF 425.00-2009] (to P.B.). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Chernoff (Fox Chase Cancer Center, Philadelphia), R. Dalla-Favera (Columbia University), V.J. Bardwell (University of Minnesota) and B. Groner (University of Frankfurt) for generously providing plasmids. Julie Millour is acknowledged for her support in setting up the ChIP technique.
REFERENCES
- 1.Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta. 2011;1813:1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 3.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–513. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 4.Benitah SA, Valeron PF, van Aelst L, Marshall CJ, Lacal JC. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim. Biophys. Acta. 2004;1705:121–132. doi: 10.1016/j.bbcan.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Boyer L, Travaglione S, Falzano L, Gauthier NC, Popoff MR, Lemichez E, Fiorentini C, Fabbri A. Rac GTPase instructs nuclear factor-кB activation by conveying the SCF complex and IkBα to the ruffling membranes. Mol. Biol. Cell. 2004;15:1124–1133. doi: 10.1091/mbc.E03-05-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matos P, Jordan P. RAC1, but not RAC1B, stimulates RELB-mediated gene transcription in colorectal cancer cells. J. Biol. Chem. 2006;281:13724–13732. doi: 10.1074/jbc.M513243200. [DOI] [PubMed] [Google Scholar]
- 7.Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran BH, Guan K. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 8.Tonozuka Y, Minoshima Y, Bao YC, Moon Y, Tsubono Y, Hatori T, Nakajima H, Nosaka T, Kawashima T, Kitamura TA. GTPase-activating protein binds STAT3 and is required for IL-6-induced STAT3 activation and for differentiation of a leukemic cell line. Blood. 2004;104:3550–3557. doi: 10.1182/blood-2004-03-1066. [DOI] [PubMed] [Google Scholar]
- 9.Kawashima T, Bao YC, Nomura Y, Moon Y, Tonozuka Y, Minoshima Y, Hatori T, Tsuchiya A, Kiyono M, Nosaka T, et al. Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors. J. Cell Biol. 2006;175:937–946. doi: 10.1083/jcb.200604073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barros P, Jordan P, Matos P. Rac1 signaling modulates BCL-6-mediated repression of gene transcription. Mol. Cell. Biol. 2009;29:4156–4166. doi: 10.1128/MCB.01813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyfert VL, Allman D, He Y, Staudt LM. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 12.Staudt LM, Dent AL, Shaffer AL, Yu X. Regulation of lymphocyte cell fate decisions and lymphomagenesis by BCL-6. Int. Rev. Immunol. 1999;18:381–403. doi: 10.3109/08830189909088490. [DOI] [PubMed] [Google Scholar]
- 13.Dent AL, Vasanwala FH, Toney LM. Regulation of gene expression by the proto-oncogene BCL-6. Crit. Rev. Oncol. Hematol. 2002;41:1–9. doi: 10.1016/s1040-8428(01)00164-0. [DOI] [PubMed] [Google Scholar]
- 14.Bajalica-Lagercrantz S, Piehl F, Farnebo F, Larsson C, Lagercrantz J. Expression of the BCL6 gene in the pre- and postnatal mouse. Biochem. Biophys. Res. Commun. 1998;247:357–360. doi: 10.1006/bbrc.1998.8551. [DOI] [PubMed] [Google Scholar]
- 15.Lin Z, Kim H, Park H, Kim Y, Cheon J, Kim I. The expression of bcl-2 and bcl-6 protein in normal and malignant transitional epithelium. Urol. Res. 2003;31:272–275. doi: 10.1007/s00240-003-0324-3. [DOI] [PubMed] [Google Scholar]
- 16.Huang YC, Hung WC, Kang WY, Chen WT, Chai CY. Expression of STAT3 and Bcl-6 oncoprotein in sodium arsenite-treated SV-40 immortalized human uroepithelial cells. Toxicol. Lett. 2007;173:57–65. doi: 10.1016/j.toxlet.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Otaki JM, Fearon DT, Yamamoto H. The proto-oncogene BCL-6 is expressed in olfactory sensory neurons. Neurosci. Res. 2005;53:189–200. doi: 10.1016/j.neures.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Kanazawa N, Moriyama M, Onizuka T, Sugawara K, Mori S. Expression of bcl-6 protein in normal skin and epidermal neoplasms. Pathol. Int. 1997;47:600–607. doi: 10.1111/j.1440-1827.1997.tb04548.x. [DOI] [PubMed] [Google Scholar]
- 19.Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L, Venkitaraman A, Wagner S. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 2003;22:5572–5578. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- 20.Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry MR, Staudt LM. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 21.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 23.Dalla-Favera R, Migliazza A, Chang CC, Niu H, Pasqualucci L, Butler M, Shen Q, Cattoretti G. Molecular pathogenesis of B cell malignancy: the role of BCL-6. Curr. Top. Microbiol. Immunol. 1999;246:257–263. doi: 10.1007/978-3-642-60162-0_32. [DOI] [PubMed] [Google Scholar]
- 24.Niu H. The proto-oncogene BCL-6 in normal and malignant B cell development. Hematol. Oncol. 2002;20:155–166. doi: 10.1002/hon.689. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 26.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 27.Matos P, Collard J, Jordan P. Tumour-related alternative-spliced Rac1b is not regulated by Rho-GDI and exhibits selective downstream signalling. J. Biol. Chem. 2003;278:50442–50448. doi: 10.1074/jbc.M308215200. [DOI] [PubMed] [Google Scholar]
- 28.Matos P, Oliveira C, Velho S, Gonçalves V, da Costa LT, Moyer MP, Seruca R, Jordan P. B-RafV600E cooperates with alternative spliced Rac1b to sustain colorectal cancer cell survival. Gastroenterology. 2008;135:899–906. doi: 10.1053/j.gastro.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 29.Fernández de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, Martino A, Nelson BH, Francis JM, Jones MC, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol. Cell. Biol. 2004;24:10058–10071. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 31.Tang TT, Dowbenko D, Jackson A, Toney L, Lewin DA, Dent AL, Lasky LA. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J. Biol. Chem. 2002;277:14255–14265. doi: 10.1074/jbc.M110901200. [DOI] [PubMed] [Google Scholar]
- 32.Meyer RD, Laz EV, Su T, Waxman DJ. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 2009;23:1914–1926. doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, Johnson KJ, Neilson LM, Liu C, Brill KL, et al. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 2010;70:1711–1721. doi: 10.1158/0008-5472.CAN-09-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calò V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Zhang Y, Glass A, Zellweger T, Gehan E, Bubendorf L, Gelmann EP, Nevalainen MT. Activation of signal transducer and activator of transcription-5 in prostate cancer predicts early recurrence. Clin. Cancer Res. 2005;11:5863–5868. doi: 10.1158/1078-0432.CCR-05-0562. [DOI] [PubMed] [Google Scholar]
- 36.Xiong H, Su WY, Liang QC, Zhang ZG, Chen HM, Du W, Chen YX, Fang JY. Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Lab. Invest. 2009;89:717–725. doi: 10.1038/labinvest.2009.11. [DOI] [PubMed] [Google Scholar]
- 37.Pelletier S, Duhamel F, Coulombe P, Popoff MR, Meloche S. Rho family GTPases are required for activation of Jak/STAT signaling by G protein-coupled receptors. Mol. Cell. Biol. 2003;23:1316–1333. doi: 10.1128/MCB.23.4.1316-1333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 40.Mermelshtein A, Gerson A, Walfisch S, Delgado B, Shechter-Maor G, Delgado J, Fich A, Gheber L. Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas. Br. J. Cancer. 2005;93:338–345. doi: 10.1038/sj.bjc.6602709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagn. Mol. Pathol. 2010;19:194–200. doi: 10.1097/PDM.0b013e3181e202f2. [DOI] [PubMed] [Google Scholar]
- 42.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell. Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Rosen DG, Yao JL, Huang J, Liu J. Expression of p14ARF, p15INK4b, p16INK4a, and DCR2 increases during prostate cancer progression. Mod. Pathol. 2006;19:1339–1343. doi: 10.1038/modpathol.3800655. [DOI] [PubMed] [Google Scholar]
- 44.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 45.Luo G, Yu-Lee L. Transcriptional inhibition by Stat5. Differential activities at growth-related versus differentiation-specific promoters. J. Biol. Chem. 1997;272:26841–26849. doi: 10.1074/jbc.272.43.26841. [DOI] [PubMed] [Google Scholar]
- 46.Nelson EA, Walker SR, Alvarez JV, Frank DA. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J. Biol. Chem. 2004;279:54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 47.Sahai E, Marshall CJ. Rho–GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.