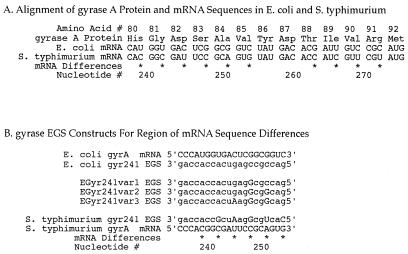
Figure 2.
Gyrase A variation between E. coli and S. typhimurium. One portion of the gyrase A molecule was studied in particular detail (see text). This portion of gyrase mRNA was used as a target for EGSs designed to assess EGSs' species specificity and nucleotide-level specificity. (A) The gyrase A protein has identical amino acid sequence in E. coli and S. typhimurium over this region. However, due to codon use variation, these two species have several differences in their respective mRNAs encoding gyrase A (*). Numbering is per EGyr sequence of Wang (16). (B) EGS oligonucleotides designed to target cleavage of this portion of mRNA at nucleotide 241. EGSs all include a 3′-acca tail and an oligonucleotide region complementary to their mRNA target Here, complemetarity is designed to be imperfect in some constructs. EGSs shown here include: EGyr241 EGS (against EGyr mRNA) and SGyr241 EGS (against SGyr mRNA); as well as three variants of the EGyr241 EGS, which differ from this EGS by step-wise introductions of one, two, or three nucleotide changes (EGyr241var1, var2, or var3 EGSs, respectively). Changes compared to EGyr241 are indicated in uppercase letters.