Abstract
We evaluated the potential of an anti–human immunodeficiency virus (HIV) Tat intrabody (intracellular antibody) to promote the survival of CD4+ cells after chimeric simian immunodeficiency virus (SIV)/HIV (SHIV) infection in rhesus macaques. Following optimization of stimulation and transduction conditions, purified CD4+ T cells were transduced with GaLV-pseudotyped retroviral vectors expressing either an anti-HIV-1 Tat or a control single-chain intrabody. Ex vivo intrabody-gene marking was highly efficient, averaging four copies per CD4+ cell. Upon reinfusion of engineered autologous CD4+ cells into two macaques, high levels of gene marking (peak of 0.6% and 6.8% of peripheral blood mononuclear cells (PBMCs) and 0.3% or 2.2% of the lymph node cells) were detected in vivo. One week post cell infusion, animals were challenged with SHIV 89.6p and the ability of the anti-HIV Tat intrabody to promote cell survival was evaluated. The frequency of genetically modified CD4+ T cells progressively decreased, concurrent with loss of CD4+ cells and elevated viral loads in both animals. However, CD4+ T cells expressing the therapeutic anti-Tat intrabody exhibited a relative survival advantage over an 8- and 21-week period compared with CD4+ cells expressing a control intrabody. In one animal, this survival benefit of anti-Tat transduced cells was associated with a reduction in viral load. Overall, these results indicate that a retrovirus-mediated anti-Tat intrabody provided significant levels of gene marking in PBMCs and peripheral tissues and increased relative survival of transduced cells in vivo.
Braun and colleagues report on an optimized procedure for the enrichment and ex vivo transduction of rhesus macaque peripheral blood CD4+ T cells with retroviral vectors expressing the anti-Tat intrabody huTat2. They report that infusion of these engineered autologous CD4+ T cells into rhesus macaques leads to detectable levels of intrabody expression in vivo and that the anti-HIV-1 Tat intrabody provides a relative survival advantage to transduced CD4+ T cells after simian–human immunodeficiency virus challenge.
Introduction
Since its discovery in 1983, human immunodeficiency virus (HIV), the causative agent of AIDS, has led to the death of close to 25 million people. Over the past 15 years, antiretroviral therapy using combinations of drugs targeting viral enzymes such reverse transcriptase, protease, and integrase or viral entry steps like attachment and fusion has greatly improved the survival of HIV-infected individuals. However, antiretroviral therapy is often associated with high toxicity, rapid emergence of resistance viral strains, and latently infected CD4+ T-cell reservoirs. Gene therapy has been proposed as a potential therapeutic modality for intracellular immunization against HIV (Baltimore, 1988). Both hematopoietic stem cells and primary CD4+ T cells may serve as the target population for expression of a variety of anti-HIV genes (Braun and Johnson, 2006). These inhibitory approaches (i.e., ribozymes, antisense, aptamers, RNAi, zinc fingers, dominant negative proteins, intrabodies, antibodies and viral decoys; Sarver et al., 1990; Jacque et al., 2002; Novina et al., 2002; Michienzi et al., 2003; Hannon and Rossi, 2004; Bahner et al., 2007; Marasco and Sui, 2007; Rossi et al., 2007; Kumar et al., 2008; Lo et al., 2008) have resulted in efficient and specific reduction of HIV replication in vitro (Brass et al., 2008; König et al., 2008; Nathans et al., 2008) and, in some cases, protection and selection of the transduced cells (Klebba et al., 2000; Humeau et al., 2004; Trobridge et al., 2009; Kimpel et al., 2010; Balazs et al., 2012).
Over the years, many viral proteins have been targeted by intrabodies, in which the antigen-binding domains of an antibody are linked and engineered for intracellular expression (Bahner et al., 2007; Marasco and Sui 2007; Lo et al., 2008). Tat is an attractive target because it has both direct and indirect effects in viral pathogenesis (Marasco et al., 1999; Romani et al., 2010) through multiple steps of the HIV-1 life cycle and modulatory effects on the host immune system. The humanized anti-Tat intrabody, huTat2, targets residues 1–20 in the N-terminal activation domain of HIV Tat and has become a leading candidate for intrabody-mediated anti-HIV gene therapy (Mhashilkar et al., 1995, 1999; Poznansky et al., 1998; Marasco et al., 1999; Poznansky et al., 1999). Previous studies have shown that retrovirus-mediated delivery of an anti-HIV-1 Tat intrabody into CD4+ T cells can lead to protective effects against laboratory and primary strains of HIV in both acutely infected and persistently infected cell lines and in transduced CD4+ mononuclear cell populations (Mhashilkar et al., 1995, 1999; Marasco et al., 1999; Poznansky et al., 1998, 1999). In studies with multiple intrabodies targeting various HIV proteins, those against Tat were the most efficient in inhibiting HIV replication both in vitro and in vivo (Mhashilkar et al., 1999).
Herein, we report optimized procedures for enrichment of rhesus macaque peripheral blood CD4+ T cells and their efficient ex vivo transduction with murine leukemia virus (MLV)-based retroviral vectors expressing the anti-Tat intrabody huTat2. Engineered autologous CD4+ T cells were administered into two rhesus macaques, and intrabody gene marking was monitored following chimeric simian immunodeficiency virus (SIV)/HIV (SHIV) infection. Our data demonstrated efficient gene transfer into CD4+ lymphocytes leading to high levels of genetically modified T cells in the peripheral circulation and secondary lymphoid tissues and to detectable levels of intrabody expression in vivo. Additionally, the anti-HIV-1 Tat intrabody provided a relative survival advantage to transduced CD4+ T cells after SHIV challenge in the two animals for a period of 2 and 5 months, which was associated with sustained CD4+ T cells counts and a reduction in viral load in one animal. Overall, upon successful transduction and optimized rates of gene transfer into HIV target cells, we found that high levels of gene marking led to long-term detection in both animals and a survival benefit of huTat2-transduced cells after SHIV infection.
Materials and Methods
Animals
Purpose-bred, Indian-origin rhesus macaques were utilized for this study. All rhesus macaques used in this study were housed at the New England Primate Research Center and maintained in accordance with institutional and federal guidelines mandated by the Animal Care and Use Committee of Harvard Medical School. Animal experiments were approved by the Harvard Medical Area Standing Committee on Animals and conducted according to the principles described in the Guide for the Care and Use of Laboratory Animals (The Institute Of Laboratory Animal Research, 1996).
Isolation of rhesus CD4+ T lymphocytes
Macaque peripheral blood mononuclear cells (PBMCs) were obtained following leukapheresis using a modification of a previously published technique (Donahue et al., 1996). Briefly, macaques were sedated with ketamine HCl (10 mg/kg intramuscularly), then intubated and anesthetized with isoflurane by inhalation and alfentanil (0.03 mg/kg/hr). Leukapheresis was performed using a COBE Spectra leukapheresis instrument (Cobe) using the Auto PBSC protocol via a femoral vein catheter and saphenous vein return. Anticoagulation was achieved using an acid citrate dextrose infusion with a CaCl2 infusion and frequent monitoring of Ca2+ levels. Pheresis was generally performed for three to four plasma volumes. After ficoll separation of leukopheresed cells, CD4+ cells were isolated by negative selection using StemSep columns (Stem Cell Technologies) according to the manufacturer's protocols. Cells were resuspended at 5×107 cells/ml in separation medium and the following antibodies were added per milliliter: 100 μl StemSep Enrichment Cocktail (anti-CD11b, CD14, CD16, CD20, CD56, and CD66e; StemCell Technologies) and 20 μl of anti-CD8 antibody complex. The mixture was incubated on ice for 30 min and EasySep™ magnetic nanoparticles at 50 μl/ml of cells were added. Following additional incubation on ice for 30 min (or room temperature for 15 min), tubes were placed in a 1-T magnetic field (Big Easy EasySep Magnet, StemCell Technologies) for 10 min and enriched CD4+ cells were decanted. To increase the yield of CD4+ cells in the second animal, the remaining cells were resuspended and then returned to the magnetic field for another 10 min before the CD4+ cells were collected again. Cells were removed from the magnet and resuspended in 8 ml phosphate-buffered saline (PBS)/2% fetal bovine serum (FBS). Small aliquots of CD4+ cells were analyzed for immunophenotyping by flow cytometric analysis.
To monitor the homing of transduced CD4+ T cells to secondary lymphoid tissues; biopsy samples of jejunum, ileum, colon, spleen, and the lymph nodes (LNs) were processed as previously described (Veazey et al., 2000). Gut biopsies and tissues were minced and treated with EDTA and type II collagenase (Sigma) during mechanical agitation. Single-cell suspensions were centrifuged over 35%–60% discontinuous Percoll gradient. To isolate lymphocytes from the spleen and axillary, inguinal, and mesenteric LNs, samples were minced and separated into single-cell suspension through a 70-μm nylon cell strainer. Small aliquots of cells were analyzed for vector frequency by real-time polymerase chain reaction (PCR) and for immunophenotyping by flow cytometric analysis.
Generation and production of the retroviral vectors
The amphotropic-pseudotyped packaging cell line Phoenix (kindly provided by Garry Nolan, Stanford) and the human osteosarcoma cell line U2OS (a kind gift from Richard Mulligan) were cultured in Dulbecco's modified Eagle's medium plus 10% FBS, 10 mM HEPES, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM L-glutamine. Each intrabody and a green fluorescent protein (GFP) marker were cloned into the MLV-based retroviral vector LZRS (also provided by Garry Nolan) and transfected into the PG13 (GaLV-pseudotyped) packaging cell line by calcium phosphate co-precipitation and washed after 24 hr. After an additional 48 hr, culture supernatant was collected, passed through a 0.45-μm filter and used to transduce Phoenix (amphotropic) packaging cell lines. Supernatant from these cells was then used to transduce PG13 packaging cells three times to increase the proviral copy number and the viral titer. Transduced PG13 producer cells were plated at limiting dilution to isolate individual clones. Multiple clones were identified, expanded, and used to measure viral titer. Dilutions of viral stocks were used to transduce U2OS cells and the percentage of transduction was determined by fluorescent activated cell sorter (FACS) analysis of GFP-expressing cells or TaqMan PCR as described below. PG13 packaging cell line containing LZRS-GFP was used as a control. Stock titers ranged from 1.0×105 to 2.6×105 transduction units (TU)/ml.
Transduction protocols
For optimization studies, T cells (2×105 cells/well or 0.1×106 cells/cm2) were prestimulated with 2.5 μg/ml αCD3 (Kawai et al., 1994) (clone: 6G12; a kind gift from Johnson Wong, MGH) and 2.5 μg/ml αCD28 (Clone: CD28.2; BD-PharMingen) antibodies linked to GAM-coated beads (polyclonal goat anti-mouse IgG Dynabeads, Invitrogen) at a 3:1 bead to cell ratio or with αCD3 and αCD28 antibodies (0.25 μg/cm2 each) linked to GAM-coated (F(ab′)2 goat anti-mouse IgG, H+L, human serum absorbed, Kirkegaard & Perry Laboratories) 24-well plates (15 μg/cm2) in RPMI-1640 medium supplemented with 10% FBS, 10 mM HEPES, 50 U/ml penicillin, 50 μg/ml streptomycin, 2 mM L-glutamine (R10), and 40 U/ml interleukin (IL)-2 (kindly provided by Dr. M. Gately, Hoffman-La Roche) for 3 days at 37°C, 5% CO2. Prestimulation of enriched CD4+ cells was performed as follows: a total of 37.5×106 cells (1×106 cells/ml or 0.5×106 cells/cm2) were resuspended in R10 plus 40 U/ml IL-2 and cultured in αCD3- and αCD28-linked GAM-coated T-75 flask at 37°C, 5% CO2 for 3 days.
To facilitate virus–cell interactions, six-well tissue culture plates were coated with the recombinant human fibronectin fragment CH-296 (Retronectin; BioWittaker) at a final concentration of 10 μg/cm2 and incubated for 2–3 hr at room temperature. Following removal of Retronectin solution, plates were coated with 2% bovine serum albumin and incubated at room temperature for 30 min. Plates were then washed with PBS prior to use. Retronectin-coated plates were preloaded with each retroviral supernatant separately over three cycles by spinning the retroviral stocks in the Retronectin-plates at 2000 rpm (900×g) for 30 min at 4°C. Transduction of the stimulated CD4+ T cells was performed for each intrabody vector by exposure to approximately a multiplicity of infection (MOI) of 2 TU per cell. For the first animal, CD4+ T cells (3×106 cells/well, 0.3125×106 cells/cm2) were seeded and cultured for 24 hr on the virus-loaded Retronectin-coated plates with additional viral supernatant, which was then removed and replaced with fresh media. For the second animal, CD4+ T cells were exposed to virus-loaded Retronectin-coated plates and were cultured for 3 days. Transduced CD4+ T cells were maintained in culture at 2×106 cells/ml R10 plus IL-2 for 1 week, before the IL-2 concentration was reduced to 20 U/ml for the final 3 days. Three to 10 days post transduction, cells were analyzed for transduction efficiency by real-time PCR.
Autologous transplantation and SHIV challenge
Six days post transduction, the transduced CD4+ T cells were prepared for autologous transplantation into rhesus macaques. The transduced cells were washed in PBS and then in 1×PBS/2% autologous serum, and a portion of cells were collected for analysis by flow cytometry and real-time PCR. The transduced cells were resuspended at 5×106 to 10×106 cells/ml in a total volume <100 ml and reinfused into the animals. Seven days post reinfusion, animals were infected intravenously with 1.5×106 copy equivalents of SHIV 89.6p (kindly provided by Keith Reimann). PBMCs, lymphoid tissue biopsies (LNs and gut), and other lymphoid tissues (tonsils and spleen) were obtained at the designated time points for phenotypic and molecular analysis.
Flow cytometric analysis
Suspensions of lymphoid tissues were resuspended in PBS with 2% mouse serum (Sigma) and stained by incubation with macaque-specific fluorescein isothiocyanate (FITC)-conjugated anti-CD20 (clone L27, BD-PharMingen), phycoerythrin (PE)-conjugated anti-CD3 antibodies (clone SP34, BD-PharMingen), allophycocyanin (APC)-conjugated anti-CD4 antibodies (clone SK3, BD-PharMingen), peridinin chlorophyll protein (PerCP)-conjugated anti-CD8 antibodies (clone SK1, BD-PharMingen), or the Simultest Control (clone X40, BD-PharMingen) for 30 min at 4°C. Naïve and memory subsets of CD4+ T cells were analyzed for expression of CXCR4 and CCR5 by staining as with PerCP-conjugated anti-CD4 antibodies, FITC-conjugated anti-CD28 antibodies (clone CD28.2, BD-PharMingen), PE-conjugated anti-CD95 antibodies (clone DX2, BD-PharMingen), and either APC-conjugated anti-CD195 antibodies (anti-CCR5, clone 3A9, BD-PharMingen), APC-conjugated anti-CD184 antibodies (anti-CXCR4, clone 12G5, BD-PharMingen), or APC fluorescence minus one negative control. The cells were washed and analyzed for the percentage of cells in each population by flow cytometry using a FACSCalibur (Becton Dickinson). When analyzed, GFP expression was measured in the FITC channel.
Molecular analysis, expression, and viral load
Genomic DNA was isolated from transduced cells, PBMCs, and tissue lymphocytes using the QIAamp Blood DNA Mini Kit (Qiagen) according to the manufacturer's protocol. For quantification of total copy number of the retroviral vectors, real-time PCR analysis was performed with universal MLV vector primers (Gerard et al., 1996) using the ABI Prism 7700 Sequence Detection System (PE Applied Biosystems). Primers and probes included: forward, CGCAACCCTGGGAGACGTCC; reverse, CGTCTCCTACCAGAACCACATATCC, and probe, 6FAM-CCGTTTTTGTGGCCCGACCTGAG-TAMRA. Reaction mixtures containing 200 ng of genomic DNA, 400 nmol each of the forward and reverse primers, and 200 nmol of the probe in 50 μl of 1×Master Mix (PE Applied Biosystems) were amplified for 40 cycles with a denaturation temperature of 94°C for 15 sec, and an annealing/elongation temperature of 64°C for 1 min. Standard curves were performed using optimized real-time PCR conditions and dilutions of HEL-GFP cells (obtained from David A. Williams), a cell line with one vector copy per cell.
To determine intrabody-specific transduction frequencies, vector-specific molecular analysis was performed with a common forward primer (5′-CTACGAGAAACACAAACTCTACGCC-3′) in Cκ sequence and the vector-specific reverse primer from the HA tag (5′-GCGTAGTCTGGGACGTCGTATGG-3′) in huTat2 or the Flag tag (5′-CTTGTCATCGTCGTCCTTGTA-3′) in A3H5. Reaction mixtures containing 200 ng of genomic DNA and 300 nM of forward and reverse primer in 25.5 μl of Syber Green PCR mixture (ABI) were amplified for 40 cycles with a denaturation temperature of 95°C for 15 sec, an annealing temperature of 68°C for the HA tag and 65°C the Flag tag for 1 min, and an elongation temperature of 72°C for 1 min. Calibration curves for the quantification of either HA or Flag gene sequences were performed using optimized real-time PCR conditions and known copy number of either purified HA or Flag retrovirus-vector DNA templates supplemented with genomic DNA from rhesus macaque PBMCs. The percent gene marking was calculated from the gene copy number based on 6.6 pg genomic DNA per cell (3×109 nt per haploid genome×average MW of dsDNA of 660 Da), or 33,000 cells per 200 ng of genomic DNA.
Samples were considered positive, but unquantifiable, when the amplification was positive, but below the lowest value on the standard curve. The relative levels of HA to Flag was given as 2−ΔCt where ΔCt is the difference between the amplification threshold for each target (Livak and Schmittgen, 2001).
To determine whether the huTat2.sFv.Cκ.HA or the A3H5.sFV.Cκ.Flag proteins were expressed and secreted from the transduced CD4+ T cells, transgene-specific enzyme-linked immunosorbent assays (ELISAs) were developed and used to analyze culture supernatants and plasma from rhesus macaque 239.96 and 308.98. The HA-specific purified IgG1 monoclonal antibody HA.11 (Clone 16B12; Covance) or the Flag-specific affinity-purified IgG1 monoclonal antibody (Clone M2, Sigma) were used as capturing antibodies. Affinity-purified goat anti-human-Cκ polyclonal antibodies (Thermo Scientific Pierce Biotechnology) were used as detection antibodies and horseradish peroxidase (HRP)-conjugated mouse anti-goat IgG H+L (ImmunoPure, absorbed with human, mouse, rabbit serum proteins, Thermo Scientific Pierce Biotechnology) was used as the secondary antibody. Specifically, the anti-HA antibody was coated onto 96-well plates (1 μg/100 μl/well), washed, and blocked with 0.5% goat serum. Supernatant and plasma samples (50 μl per well) were bound for 1 hr at room temperature, followed by six washes. The goat anti-human Cκ IgG was used at 4 μg/ml and the HRP-conjugated secondary antibody was used at 1:10,000 dilution. The substrate 3,3′,5,5′-tetramethylbenzidine (TMB, KPL Inc.) was converted by HRP to a colored precipitate and the optical density (OD) was assessed at 450 nm per manufacturer's instruction. The background OD was subtracted from the sample OD to provide relative values.
Quantification of SHIV plasma RNA was performed by bDNA assays as previously described (Chiron/Bayer Diagnostic) (Sodora et al., 1998). The lower limit of detection of this assay in this study was 400 copies per milliliter.
Results
In vitro transduction of CD4+ lymphocytes
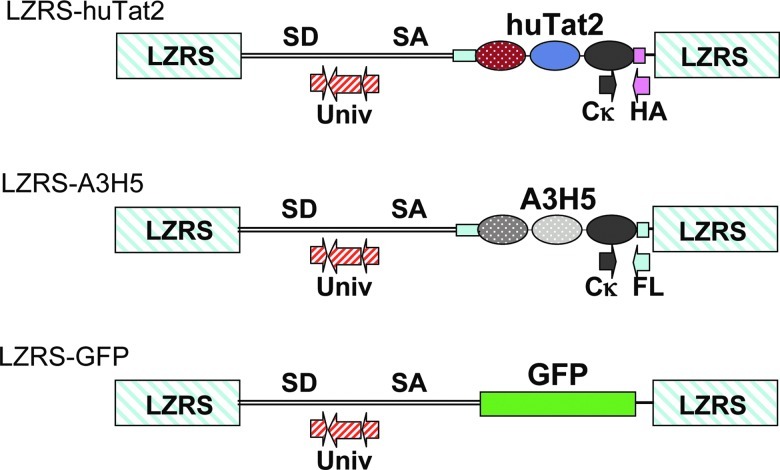
In order to evaluate the efficacy of huTat2 intrabody in vivo, the therapeutic vector LZRS-huTat2 (Fig. 1) was generated to express huTat2, a human intrabody that targets the N-terminal activation domain of HIV Tat. huTat2 intrabody consists of a single chain fragment (scFv) that is fused in frame at its C-terminal with the human immunoglobulin κ light chain constant domain (Cκ) for increased stability (Palmer et al., 2006). In such a format, huTat2 protects transduced cells from HIV challenge (Mhashilkar et al., 1999). An human IgG1 leader sequence was fused to the N-terminus of huTat2, leading to higher levels of intrabody expression due to the presence of endogenous chaperones in the endoplasmic reticulum (ER) that assist with intrabody folding. As a control for intrabody-mediated protection against viral infection, the retroviral vector LZRS-A3H5 (Fig. 1) was generated to express the A3H5 intrabody. A3H5 targets the 34 amino acid transformation effector site-1 motif of the cytoplasmic tail of the oncogenic latent membrane protein 1 (LMP1) of Epstein Barr virus (Gennari et al., 2004). MLV-based retroviral vectors, based on the LZRS system (Kinsella and Nolan, 1996), were shuttle-packaged through Phoenix (amphotropic) packaging cells into the GaLV-pseudotyped packaging cells lines PG13 to generate high-titer producer cell lines with titers ranging from 1.0×105 to 2.6×105 TU/ml of cell supernatant. In this study, we used these retroviral vectors to transduce a large number of CD4+ T cells for adoptive transfer into rhesus macaques and then examined whether the anti-huTat2 intrabody would protect CD4+ T cells that were challenged with SHIV.
FIG. 1.
Schematic diagram of MLV-based vectors. The MLV-based gamma-retroviral vector LZRS was used to express either green fluorescent protein (GFP), the therapeutic single chain fragment (scFv) intrabody (Leader.huTat2.Cκ.HA), or the control scFv intrabody (Leader.A3H5.Cκ.Flag); the intrabody fusion proteins used the human IgG leader to direct expression to the endoplasmic reticulum, the common kappa region (Cκ) to increase stability, and either the human influenza hemagglutinin (HA) protein (YPYDVPDYA) or FLAG epitope (DYKDDDDK) to tag protein expression. The universal primers and probe (Univ, striped arrows), located in the Psi packaging region of each vector, monitor total transduction frequencies. The vector-specific forward primer was located in the Cκ sequence downstream of the intrabody gene (grey arrow) and the vector-specific reverse primers were located in either the HA or FLAG sequences (grey arrow). Color images available online at www.liebertonline.com/hum
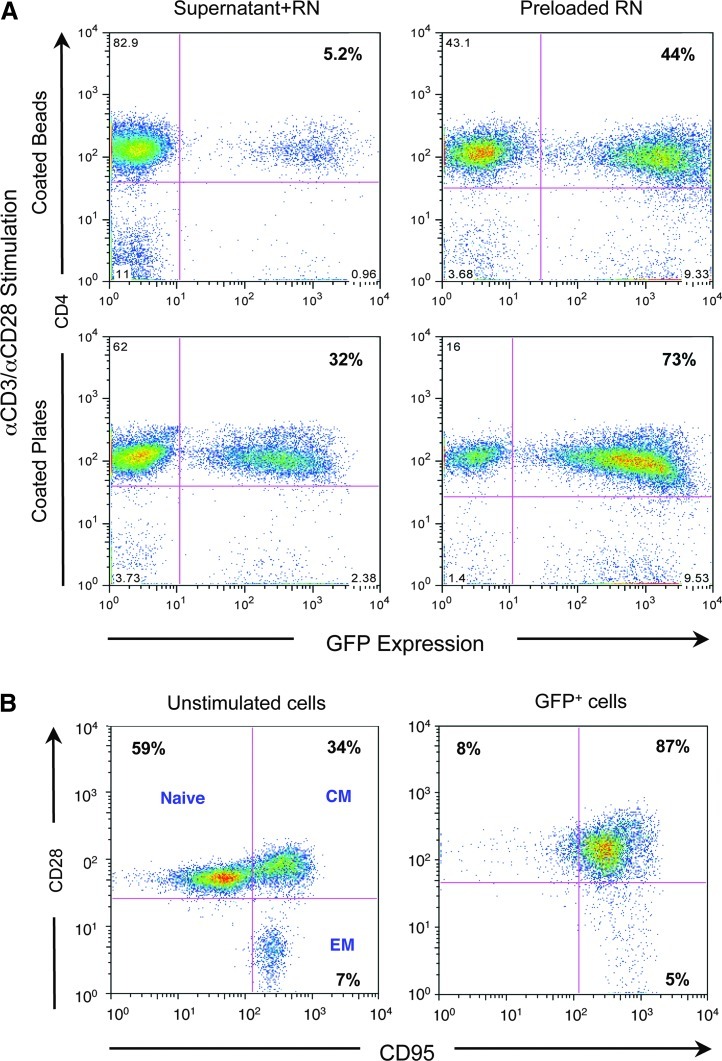
Initial studies focused on optimization of transduction of macaque CD4+ T cells. Previous studies had compared stimulation and transduction using concanavalin A (ConA), phytohemaglutinin (PHA), or the clinically relevant anti-CD3/anti-CD28 activation protocols (Zhang et al., 2003). Here, a number of parameters were analyzed to optimize efficient gene delivery to CD4+ T cells using an MLV-based retroviral vector expressing GFP (Fig. 1) and a vector expressing an intrabody without GFP. To compare various clinically relevant activation protocols, rhesus PBMCs were stimulated through the T-cell receptor (CD3) and CD28 costimulatory receptor using anti-CD3/anti-CD28 antibodies either coated to microbeads or directly to tissue culture plates and supplemented with IL-2. Both stimulation protocols activated CD4+ T cells as determined by changes in their phenotype (data not shown). However, proliferation and transduction of CD4+ T cells stimulated with antibodies attached directly to the tissue culture plates (Fig. 2A, lower panels) was significantly more efficient than antibodies attached to the beads (Fig. 2A, upper panels), at least under these conditions. The virus-binding domain of the recombinant human fibronectin fragment CH-296 (Retronectin) has been shown to enhance transduction of CD4+ T cells (Pollok et al., 1998), partially through colocalization of the viral particle with the target cell (Hanenberg et al., 1997). To determine the feasibility of preloading viral particles to the plate, Retronectin-coated plates were used for viral absorption prior to transduction of the activated cells. The efficiency of transduction was compared for viral particles, either preloaded onto Retronectin or contained in the transduction of the CD4+ T cell supernatant. After activation of CD4+ T cells, transduction of the CD4+ T cells was more efficient when the viral particles were preloaded to the Retronectin-coated plates (Fig. 2A, right panels vs. left panels).
FIG. 2.
In vitro optimization for transduction of macaque CD4+ T cells. (A) Rhesus macaque peripheral blood mononuclear cells (PBMCs) were stimulated for 3 days with anti-CD3/anti-CD28 antibodies either immobilized to beads (top row) or to the plate (bottom row) and transduced overnight using an estimated multiplicity of infection (MOI) of 2 transduction units (TU)/cell either directly with supernatants from GaLV-pseudotyped GFP-or intrabody-expressing retroviral vectors (left column) or with supernatants preloaded on Retronectin-coated plates (right column) by spinning at 900×g for 30 min. CD4+ T cells were analyzed for GFP expression after 3 days by flow cytometry. (B) Fresh (left panel) or stimulated (right panel) CD4+ T cells were transduced to express GFP and assessed for expression of CD28 and CD95, molecules which permit categorization of T cells into naïve, central memory (CM), and effector memory (EM) populations.
Molecular analysis of proviral integration using real-time PCR confirmed the increased transduction demonstrated by GFP expression. We also extended these molecular findings using the control vector LZRS-A3H5 (data not shown). We have observed similar levels of transduction with both GaLV and amphotropic pseudotyped vectors and with both human and rhesus cells (data not shown).
Finally, to assess the phenotype of transduced cells after stimulation and transduction, fresh CD4+ T cells were compared with transduced CD4+ cells for expression of the naïve and memory markers CD28 and CD95 (Pitcher et al., 2002). As shown in Fig. 2B, prior to stimulation, fresh CD4+ cells contained a mixture of naïve, central memory, and effector memory populations. In contrast and as expected after activation, the population of transduced CD4+ cells was mostly CD28+ and CD95+ central memory cells. In addition, while the naïve cells expressed more CXCR4, the central memory cells expressed both CXCR4 and CCR5, before and after stimulation (data not shown). More than 87% of the CD4+ cells were transduced under these experimental conditions (data not shown). These data indicate that CD4+ T cells activated with signals that mimic natural stimulation are transduced at high efficiency, particularly when the viral particles are bound to Retronectin, and these transduced cells lose their naïve phenotype and become central memory cells.
Ex vivo gene marking and adoptive transfer of transduced CD4+ T cells
Next, large-scale ex vivo transduction of rhesus macaque CD4+ cells was conducted. Both the therapeutic LZRS-huTat2 and control LZRS-A3H5 vectors were included to determine whether the anti-Tat intrabody could promote an in vivo survival advantage to CD4+ T cells relative to cells expressing the control intrabody following viral challenge. To accomplish this aim, PBMCs were collected from two individual animals (referred to as 306.98 and 239.96). CD4+ T cells were enriched and then transduced ex vivo with retroviral vectors expressing either the huTat2 or control intrabodies, and reinfused back into the animal. Table 1 summarizes the clinical characteristics of the two rhesus macaques and of the transduced CD4+ cells used for adoptive transfer. In these studies, the two rhesus macaques were closely matched for age, sex, weight, total cell recovery, in vitro gene transfer efficiency, and the number of transduced cells that were reinfused. Optimization experiments indicated that two rounds of CD4+ cell separation increased the yield without increasing the contamination with other cell types, and this was implemented in animal 239.96 (data not shown). Following leukapheresis, density-gradient centrifugation, and CD4+ T-cell enrichment (depleted of CD8, B and natural killer [NK] cells), more than 1×108 CD4+ cells were obtained from each animal. Following stimulation of cells with anti-CD3 and anti-CD28 antibodies (plate bound) plus 40 U/ml IL-2 for 3 days, CD4+ T cells were transduced with either huTat2 or the control A3H5 vectors at an MOI equivalent to 2 TU/cell either absorbed by Retronectin and/or provided in the supernatant. After 1 week in ex vivo culture, the IL-2 concentration was reduced to 20 U/ml for 3 more days. Then, transduced cells were pooled and infused back into each animal (Table 1). Initial proviral copy number in the transduced cell population was determined by real-time PCR for each vector prior to reinfusion of cells (Table 1). Overall, these results demonstrate highly efficient retrovirus-mediated gene transfer into clinical-scale quantities of purified CD4+ T cells using a single exposure to a relatively low MOI transduction using Retronectin.
Table 1.
Clinical Parameters of Animals and Assessment CD4+ T Cells Transduction.
| |
|
|
|
Transduced cells reinfused (×106) |
Vector copy (no. per cell) |
||
|---|---|---|---|---|---|---|---|
| Animal | Weight (kg) | Leukapheresis yield (×106 PBMC) | Purified CD4+ T cells (×106) | hu Tat2 | A3H5 | huTat2 | A3H5 |
| 306–98 | 13.5 | 825 | 137 | 37 | 117 | 3.3 | 1.8 |
| 239–96 | 13 | 450 | 116 | 95 | 115 | 2 | 8.9 |
Assessment of initial levels of in vivo gene marking
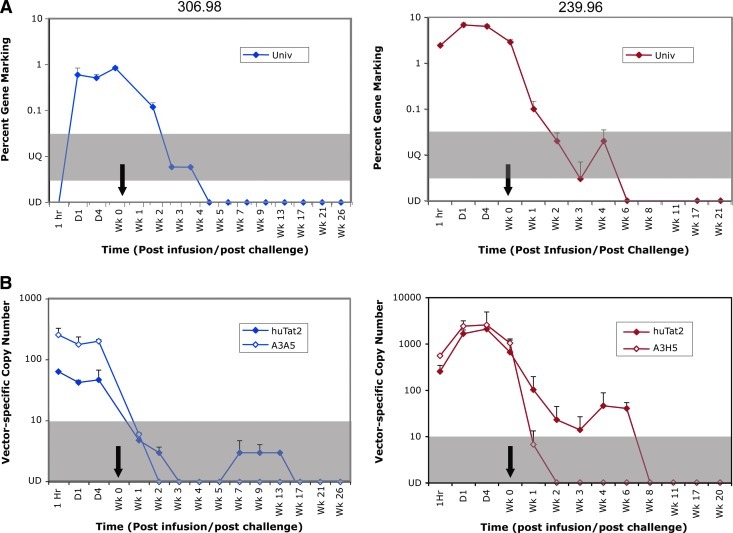
To determine the total level of in vivo gene marking in the CD4+ T cells transduced with the MLV-based retroviral vectors, PBMCs from serial time points were analyzed by real-time PCR for vector copy number using a set of universal primers (Univ) located on the Psi packaging region that is common to all the MLV vectors (Fig. 1). As shown in Fig. 3A (right panel), PBMCs isolated from animal 239.96 exhibited peak levels of overall gene transfer close to 10% of PBMCs prior to SHIV challenge. The initial frequency of gene marking in animal 306.98 was 0.7% of PBMCs (Fig. 3A, left panel).
FIG. 3.
Molecular analysis of in vivo gene marking. (A) Overall gene marking in PBMCs, collected from rhesus macaques (306.98 left panel; 239.96 right panel) at the designated time points, was monitored by real-time PCR for the percentage of cells containing retroviral vectors using the Universal primers and probe positioned within the MLV vector backbone. (B) Samples were analyzed for transduction of intrabody-specific vectors at the designated time points by real-time PCR, using primers positioned downstream of the scFv genes in the Cκ region and either the HA (huTat2, filled symbols) or Flag (A3H5, opened symbols) sequences. Values are expressed as number of copies per 200 ng genomic DNA. For all plots, 10 copies per 200 ng genomic DNA is equivalent to 0.03% gene marking. The gray bar defines the area that is positive, but below the lowest value on the standard curve. These samples were considered unquantifiable (UQ). Undetectable (UD) samples are plotted on the x-axis. The black arrows designate the time of chimeric simian immunodeficiency virus (SIV)/human immunodeficiency virus (SHIV) 89.6p challenge (1 week post infusion). Color images available online at www.liebertonline.com/hum
To determine the relative levels of therapeutic and control vectors and to assess the relative survival advantage of CD4+ cells following SHIV infection, vector-specific primers (Fig. 1) were also used to quantify the huTat2 and the A3H5 vectors in serial PBMC samples by real-time PCR. As demonstrated for animal 306.96 (Fig. 3B, left panel), prior to SHIV challenge both vectors had frequencies between 50 and 250 copies per 200 ng of genomic DNA (or approximately 1% of the PBMCs). For animal 239.96 (Fig. 3B, right panel), the frequencies of each vector peaked at 2123 and 2623 copies per 200 ng of genomic DNA (or 6.4% and 7.8% of PBMCs) for the huTat2 and A3H5 vectors, respectively. Together, these data demonstrate adoptive transfer of transduced cells leads to relatively high levels of vector-transduced cells in both animals prior to SHIV infection.
Plasma viremia following SHIV challenge
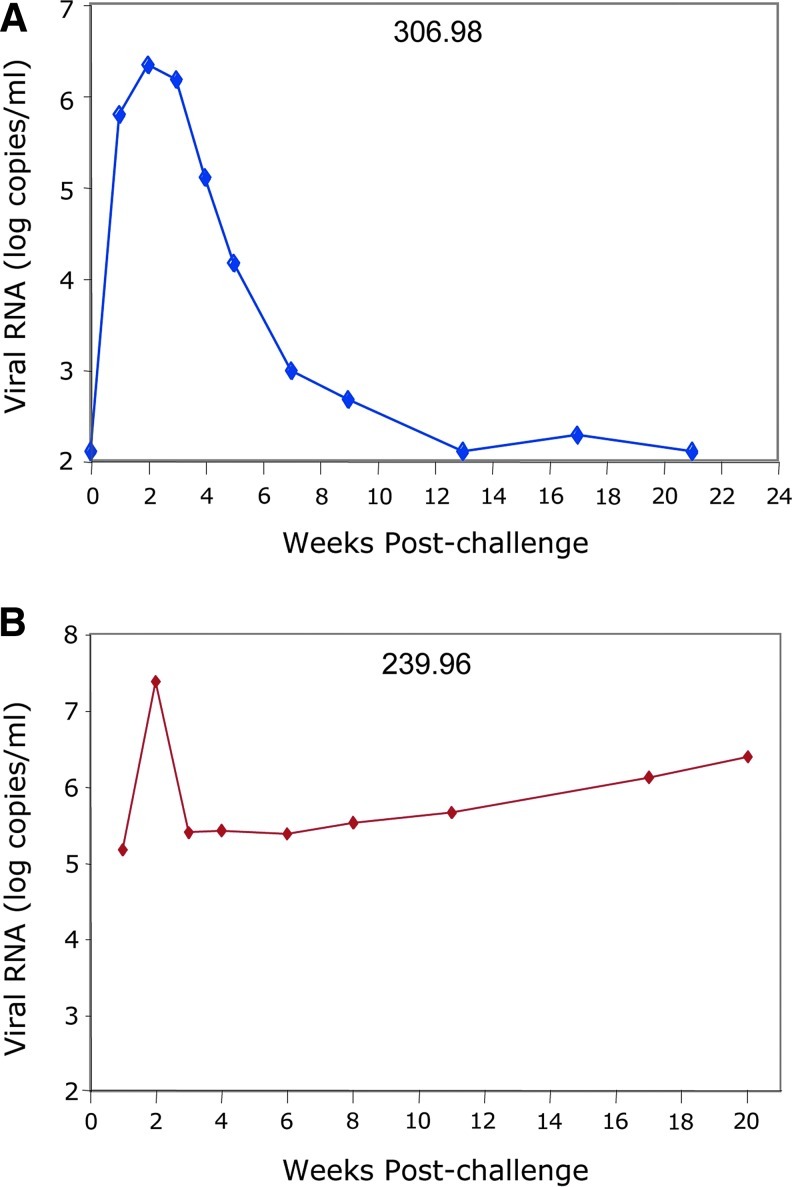
To evaluate the potential therapeutic benefit of anti-Tat intrabody engineered CD4+ cells, animals were challenged intravenously with SHIV 89.6p (1.5×106 copy equivalents intravenously) 7 days after adoptive transfer of the transduced CD4+ T cells. Serial plasma samples were collected post challenge and analyzed for levels of SHIV RNA. As expected, viremia peaked in animals 306.98 and 239.96, respectively, at 2.1×106 and 2.3×107 copies/ml 2 weeks post SHIV challenge (Fig. 4). Viral replication in animal 306.98 subsequently decreased below the level of detection (approximately 103 copies/ml), while viral replication in animal 239.96 decreased but stabilized at its set point (approximately 105 viral copies/ml) and then progressively increased over time (Fig. 4, bottom panel). The control of viremia in macaque 306.98 occurred despite lower levels of initial gene marking compared with macaque 239.96.
FIG. 4.
Plasma viremia following SHIV challenge. At week 1 after infusion of transduced CD4+ T cells, animals were infected with SHIV 89.6p and plasma was collected at the designated time points. Viral loads were quantified using the Bayer branched chain DNA assay and are presented for (A) 306.98 and (B) 239.96 as log copy number per milliliter. Color images available online at www.liebertonline.com/hum
CD4+ T cells in PBMC and tissues following SHIV challenge
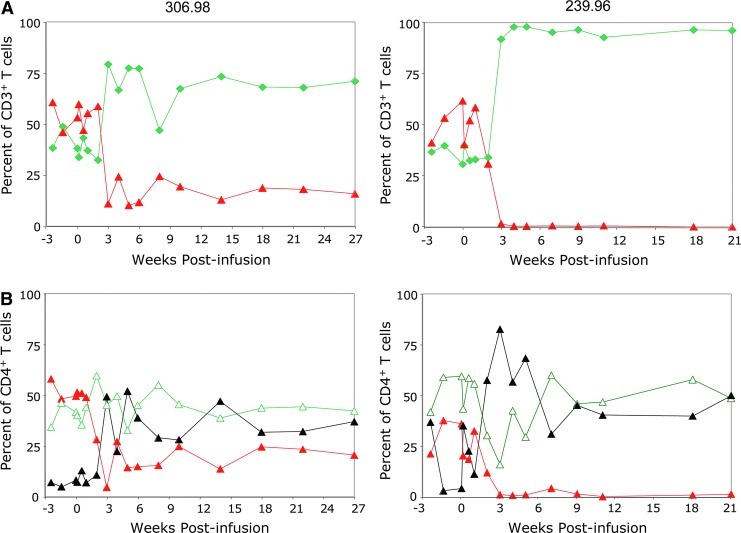
To assess the effects of viral replication on key indicators of disease progression, the frequencies of CD4+ and CD8+ T cells and of memory subsets in PBMC and selected secondary lymphoid tissues (LNs and gut) were determined at serial time points following viral infection. For both animals, the baseline percentage of CD4+ T cells in PBMC was 60%. Within 2 weeks of SHIV challenge, CD4+ T cells sharply and irreversibly declined, reaching levels of approximately 10%–20% in 306.98 and less than 1% in 239.96 (Fig. 5A), resulting in an inverted ratio of CD4+ to CD8+ cells characteristic of HIV infection. Although all T-cell subpopulations were affected, the CD28+/CD95low naïve subpopulation declined more than the memory subpopulations and with corresponding increases in the CD28−/CD95+ effector memory subpopulation (Fig. 5B), as was observed previously in macaques infected with CXCR4-tropic pathogenic SHIV strains (Nishimura et al., 2005).
FIG. 5.
The percentage and phenotypic characterization of CD4+ T cells in PBMCs following SHIV challenge. (A) PBMCs, collected from rhesus macaques (306.98 left panel; 239.96 right panel) at the designated time points post reinfusion, were stained with anti-CD3, anti-CD4, anti-CD8, and anti-CD20 antibodies to determine the percentage of CD4+ (red/gray triangle) and CD8+ (green diamond) T cells by flow cytometry. Animals were challenged with SHIV 89.6p 1 week post reinfusion. (B) PBMCs were stained with anti-CD4, anti-CD28, and anti-CD95 antibodies at serial time points post SHIV infection, and the percentage of naïve (red/gray triangle), central memory (green open triangle), and effector memory (black triangles) cells determined by flow cytometry. Color images available online at www.liebertonline.com/hum
Similarly, CD4+ T cells declined in secondary lymphoid tissues of both animals after SHIV challenge (data not shown), although the decline in CD4+ T cells in the LNs of 306.98 was not as extensive as in PBMCs. Additionally, naïve CD4+ T cells were still abundant in LNs compared to the same time points in PBMCs with essentially no effector memory cells. In 239.96, which had uncontrolled viremia, CD4+ T cells and especially naïve subpopulations declined to less than 1% of LNs (data not shown). Overall, the stable levels of CD4+ T cells in 306.98 and complete loss of CD4+ T cells in 239.96 correlated to the relatively low and controlled chronic viral loads in 306.98 and the high viral loads in 239.96.
Assessment of in vivo gene marking following SHIV challenge
To determine the total level of in vivo gene marking and the relative levels of the therapeutic and the control vectors following SHIV infection, PBMCs and other lymphoid tissues were collected at serial time points and analyzed for gene marking by real-time PCR. Initial levels of overall gene marking declined in PBMCs during acute infection as the CD4+ T cells declined following SHIV challenge (Fig. 3A), becoming undetectable at week 4 and 6 for animal 306.98 and 239.96, respectively (Fig. 3A).
For the therapeutic and control vectors, the frequency of the A3H5 control vector and the huTat2 therapeutic vector declined after SHIV challenge; however, the decline in the control vector was more rapid than the therapeutic huTat2 vector in both animals (Fig. 3B). Additionally, the therapeutic vector subsequently increased in frequency transiently post challenge in both animals (Fig. 3B). Thus, vector-specific analysis of the PBMC confirms the findings in the total gene marking analysis—an overall decline during acute infection—and reveals the subsequent transient preferential expansion of the huTat2-transduced cells.
Gene marking in lymphoid tissues
Lymphocytes normally circulate in and out of tissues and the blood. Thus, levels of transduced cells in the blood may change by migrating to secondary lymphoid tissues. To assess the total level of gene marking in tissues, we collected LNs, gastrointestinal tissues, tonsils, and spleen samples, and the isolated lymphocytes were analyzed by real-time PCR. In LNs of both animals, the total gene marking at day 7 post-reinfusion was slightly reduced from the marking in the PBMCs (2.2% of cells in LN tissue for animal 239.96, and 0.3% for animal 306.98) using the universal MLV (Table 2). In contrast, gene marking was detected at only low levels in the colon of 239.96 after 1 week, indicating less efficient homing of transduced cells to mucosal lymphoid tissues. In both animals, total gene marking in the tissues decreased after SHIV challenge following the levels in PBMCs. For animal 306.98, vector sequences were still detectable in LNs 26 weeks post viral challenge using the universal MLV primers, while only barely detectable in LN cells for 239.96 6 weeks post viral challenge. In other tissues, the universal MLV real-time PCR was also positive in the spleen of animal 306.98 at week 26 post viral challenge, while other tissues (inguinal, mesenteric, and tonsil LNs, and duodenum, jejunum, and ileum gastrointestinal samples) collected 6 months post SHIV challenge were undetectable. These results demonstrate long-term adoptive transfer of transduced CD4+ T cells even 5–6 months after a single infusion of transduced cells. Additionally, similar levels of gene marking in the LNs, in contrast to reduced gene marking in the gastrointestinal tissue, indicate a more efficient exchange of transduced lymphocytes with the LNs than with gut tissues.
Table 2.
Total Gene Markinga in Secondary Lymphoid Tissues
| 306.98 | 239.96 | |
|---|---|---|
| Lymph Node | ||
| Wk 0 | –b | NDc |
| Wk 1 | 0.3 | 2.2 |
| Wk 6 | – | 0.03 |
| Sacd | 0.007 | ND |
| Gastrointestinal | ||
| Wk 0 | – | ND |
| Wk 1 | – | 0.03 |
| WK 6 | – | ND |
| Sac | ND | ND |
| Spleen | ||
| Sac | 0.03 | ND |
Percent positive with the Universal Primer and Probe.
Samples were not collected.
ND is not detectable.
Mm 306.98 was sacrificed at week 27 and 239.96 at week 21. Lymphoid tissues collected at sacrifice were inguinal, mesenteric and tonsil LNs; spleen; jejunum, duodenum, ileum gastrointestinal samples.
To determine the relative levels of the therapeutic and the control vectors, cells from tissues (LNs, gut, spleen, ileum, duodenum, and jejunum) of macaque 239.96 were analyzed by vector-specific real-time PCR (Table 3). For cells from the LNs, initial levels of vector-specific gene marking slightly favored the control A3H5 vector over the huTat2, reaching 907 and 608 copies per 200 ng of lymphocytes genomic DNA, respectively, with essentially equivalent marking in the PBMCs. Six weeks after SHIV challenge, huTat2 sequences were detected in the LNs indicating better survival of the huTat2 vector in vivo. The huTat2-specific amplicons were also detected at much higher frequency in multiple LN samples (inguinal, mesenteric, auxiliary) at the time of sacrifice than the control A3H5 amplicon. In samples from the gastrointestinal tract, gene marking was low in most samples at most time points, which would suggest inefficient homing to gastrointestinal sites. The huTat2 vector was also detected in the spleen at the final time point. These results confirm the long-term selective survival of huTat2-transduced CD4+ T cells in secondary lymphoid tissues of animal 239.96 through the terminal week 26 of our study.
Table 3.
Vector-Specific Markinga in Secondary Lymphoid Tissues from 239.96
| Lymphoid Tissues | huTat2-HA | A3H5-Flag | Relative Ratiob |
|---|---|---|---|
| Lymph nodes | |||
| Week 0 | NDc | ND | 1 |
| Week 1 | 608 | 907 | 0.2 |
| Week 6 | 26 | ND | 7.3 |
| Auxiliary -Sac | 9 | 3 | 3.0 |
| Inginual -Sac | 7 | 1 | 2.3 |
| Mesenteric -Sac | 26 | 2 | 10.2 |
| Spleen | 4 | ND | 2.9 |
| Gastrointestinal | |||
| Week 0 | ND | ND | 0.7 |
| Week 1 | ND | ND | 1 |
| Week 6 | ND | 1 | 1 |
| Week 21 | ND | 1 | 0.7 |
| Jejunum | ND | 10 | 0.7 |
| Duodenum | 3.4 | ND | 2.4 |
| Ileum | ND | ND | 1 |
Vector copies per 200 ng genomic DNA.
Relative ratio is calculated as 2−ΔCt.
ND is not detectable.
Expression of intrabodies in vivo
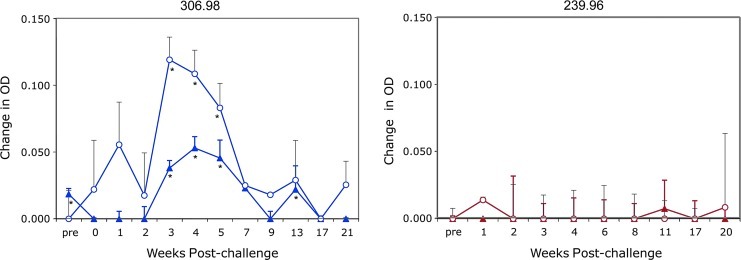
Since the intrabodies have the human IgG leader sequence and are processed naturally through the ER for secretion, we developed an ELISA to detect either the therapeutic huTat2.Cκ.HA and control A3H5.Cκ.Flag fusion proteins. In plasma from 306.98 following adoptive transfer of transduced cells, the production of both the huTat2 and A3H5 intrabodies was detected at weeks 3–5 (Fig. 6) and the huTat2 at week 13. Expression of the intrabodies in the plasma of 239.96 was not significant (p>0.05). The higher in vivo levels of intrabody expression correlated with the higher levels of intrabody detected in the conditioned media of transduced cells from 306.98 (data not shown). Importantly, these results demonstrate not only efficient transduction and efficient maintenance of the transduced CD4+ T cells in animals after adoptive transfer and after viral challenge, but also detectable levels of in vivo expression of the secreted intrabodies.
FIG. 6.
Expression of the huTat2 and A3H5 intrabodies in plasma following SHIV challenge. Plasma from 306.98 (left panel) and 239.96 (right panel) was collected at serial time points post SHIV infection and analyzed for intrabody secretion by enzyme-linked immunosorbent assay (ELISA). The therapeutic huTat2 fusion protein (filled triangles) was Cκ.HA-tagged and the control A3H5 fusion protein (open circles) was Cκ.Flag-tagged. Using a HA- or Flag-specific antibody to bind the protein to the plate, an anti-Cκ antibody was used for specific detection of each intrabody. Since a quantitative standard curve is unavailable, the background optical density (OD) was subtracted from each sample and graphed as the change in OD. The standard deviation for the background was 0.002 units in the HA assay and 0.015 in the Flag assay. *Denotes those samples with a statistically significant (p>0.05) difference OD compared to background. Color images available online at www.liebertonline.com/hum
Survival advantage of HuTat2-transduced CD4+ T cells after SHIV challenge
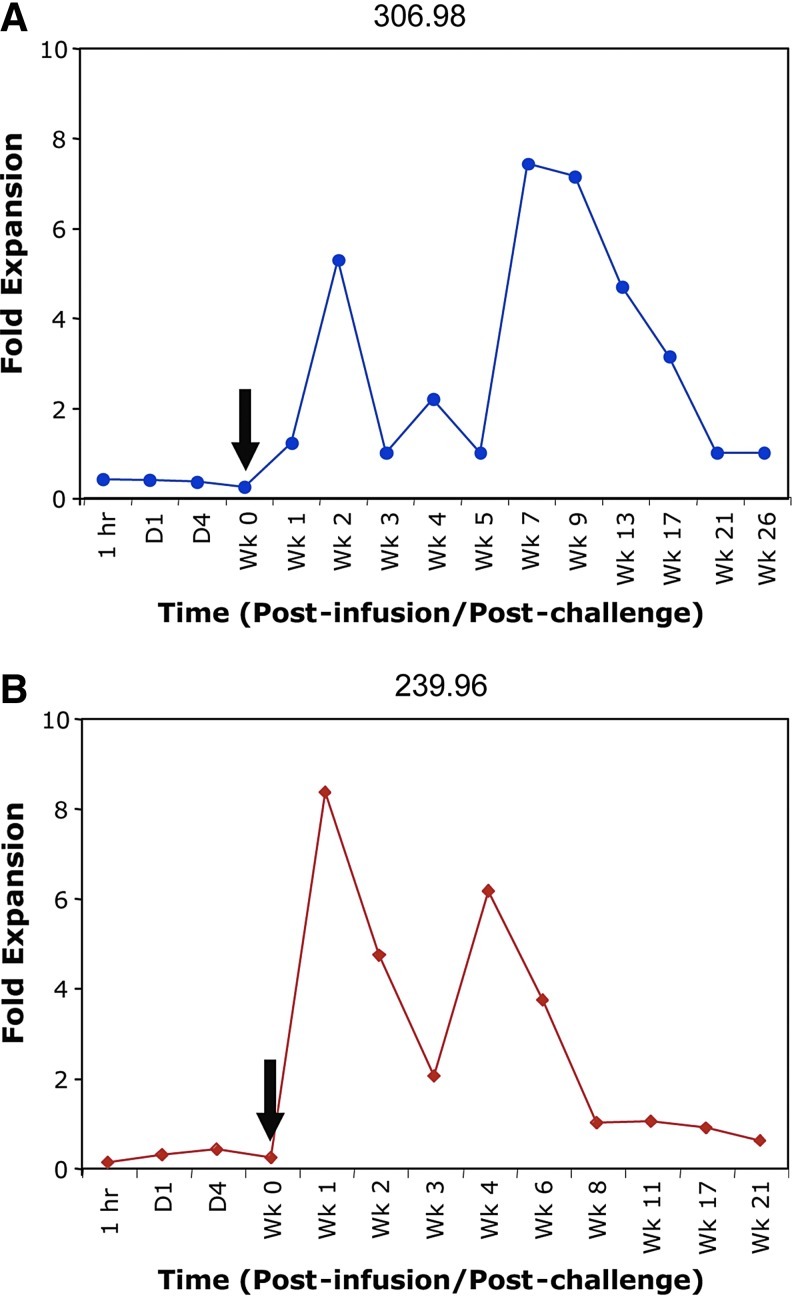
Finally, our results demonstrated long-term survival of transduced CD4+ T cells in multiple lymphoid tissues and expression of the intrabodies in the plasma after SHIV challenge. To assess whether expression of the anti-Tat intrabody in CD4+ T cells promotes a survival advantage to transduced cells, we compared the relative frequency of the therapeutic huTat2 vector to the control A3H5 vector by calculating the exponential change with the difference in Ct values in real-time PCR (Fig. 7) (Livak and Schmittgen, 2001). Prior to SHIV challenge (day 0 until day 7), the relative levels of the huTat2 and A3H5 vectors in PBMCs were constant (ranging between 0.1 and 0.4 units) with more copies of the A3H5 and a relative ratio less than 1. During the acute phase of viral infection and during very strong depletion of the CD4+ T cells (starting at week 1 post viral challenge), the relative levels of the huTat2 peaked at a ratio of 5.3- and 8.3-fold for animal 306.98 and 239.96, respectively, before returning to baseline. These data indicate increased survival of the huTat2 vector transduced cells after challenge. Interestingly, the ratio of huTat2 to A3H5 vector sequences increased again between weeks 4 and 7, corresponding to observed increases in the total gene marking. The increased ratio of huTat2 was maintained in animal 306.98 past week 17. These data show that huTat2-transduced cells in blood have a preferential survival advantage, as compared with A3H5-transduced cells.
FIG. 7.
Survival advantage of huTat2-transduced CD4+ T cells after SHIV challenge. The ratio of the therapeutic huTat2 vector to the control A3H5 vector in (A) 306.98 and (B) 239.96 was calculated using the difference in the amplification threshold between the huTat2 and the A3H5 vector-specific real-time PCR. A ratio greater than one implies more of the huTat2 intrabody vector than the A3H5 vector. The black arrows designate the time of SHIV89.6P challenge (1 week post infusion). Color images available online at www.liebertonline.com/hum
Analysis of vector-specific sequences revealed long-term engraftment of transduced CD4+ T cells in LNs and gastrointestinal tissues (Table 3). A comparison of the relative survival of the therapeutic huTat2 vector with the control A3H5 vector in secondary lymphoid tissues from 239.96 found 35-fold expansion in the relative levels of the huTat2—from 0.2 at week 1 (prior to SHIV challenge) to 7.3 at week 6 (post challenge). The increased relative ratio continued in various LN samples past 21 weeks in 239.96 (Table 3) and in 306.98 (data not shown). In gastrointestinal tissues at sacrifice, the levels of gene marking were low but detectable, and the ratio of the huTat2 and A3H5 were not substantially increased. These results in lymphoid tissues reaffirm the increased relative survival of huTat2 vector seen in PBMCs.
Discussion
In this report, the ability of a retrovirus-expressed huTat2 intrabody (Mhashilkar et al., 1995) to protect CD4+ T cells and promote a selective survival advantage in transduced cells after SHIV challenge was evaluated in the rhesus macaque animal model. An optimized ex vivo transduction protocol was established and its use resulted in high levels of gene transfer into the purified CD4+ T cells with the MLV-based retroviral vector and efficient gene marking in PBMCs and secondary lymphoid tissues following reinfusion. After animals were challenged with SHIV, viral replication led to a substantial decrease in CD4+ T cells in both animals, with corresponding decrease in gene marking. Nevertheless, gene marking was detectable in both animals even 5–6 months after SHIV challenge with relatively more abundant levels of huTat2 compared with the control A3H5 vector, indicating positive selection of the huTat2-transduced cells. In addition, these results were associated with decreased viral loads and steady CD4+ cell counts in animal 306.98. Overall, we conclude that a retrovirus-mediated delivery of huTat2 provided high levels of gene marking, in both PBMCs and peripheral tissues, and relative selection of huTat2-transduced cells in vivo through approximately 2 and 5 months for animals 239.96 and 306.98, respectively.
The HIV-1 transactivating factor Tat is an important therapeutic target for HIV therapy due to its multiple roles in the pathogenesis of AIDS. Originally, Tat was identified as a potent activator of viral transcription through association with numerous cellular proteins involved in the host transcription including cyclin T1, RNA polymerase II, protein kinase R, p300/CBP, and P/CAF (Harrich et al., 2006) and by direct interaction with viral proteins including reverse transcriptase (Apolloni et al., 2003). Although normally processed to the nucleus, Tat protein is also secreted by infected cells and, due to its structure, taken up by neighboring cells, causing bystander cell activation and permissiveness to HIV-1 infection (Peruzzi, 2006). This T-cell activation can lead to accelerated cell turnover and increased apoptosis (Galati et al., 2002).
Several reports have demonstrated efficient use of anti-HIV intrabodies, in the form of single-chain fragments (scFv), that provide protection of T-cell lines and primary CD4+ cells against viral replication and facilitate selective survival advantage to transduced cells (Swan et al., 2006). Additionally, intrabodies targeting Tat functions have been shown to be highly effective in inhibiting HIV replication both in vitro and in vivo (Mhashilkar et al., 1995; Poznansky et al., 1998, 1999; Marasco et al., 1999; Bai et al., 2003). To inhibit both the intracellular and extracellular effects of Tat, we expressed the huTat2 intrabody with the N-terminal IgG leader and a C-terminal human immunoglobulin Cκ from an MLV-based retroviral vector in CD4+ T cells. The IgG leader directs expression of the huTat2 to the ER, where the intrabody can be secreted or retrograde transported through the nuclear membrane into the nucleus by direct retrograde and/or indirect cytoplasmic import mechanism(s). We have previously demonstrated that intrabody folding efficiency and protein stability are much greater when they are expressed in the ER as compared with the cytoplasm (data not shown). As evident here, transduction and expression of the intrabodies in transduced CD4+ T cells led to detectable levels of protein in the plasma. Other adoptive T-cell transduction studies have not shown protein expression but rather have only shown RNA expression after in vitro stimulation (Bunnell et al., 1997). We hypothesized that expression of the huTat2 intrabody in transduced cells will inhibit Tat functions, such as enhancement of T-cell activation or transactivation (Peruzzi, 2006), protect transduced cells from viral pathogenesis, and provide a selective survival advantage. In addition, the secreted huTat2 intrabody can inhibit Tat protein uptake by bystander and neighboring CD4+ T cells and inhibit their permissiveness to SHIV infection.
The study herein also demonstrates efficient gene marking and detectable expression of intrabody genes in vivo through optimized procedures for the retrovirus-mediated transduction of purified CD4+ T cells. The in vitro gene marking obtained during the large-scale transduction protocol was higher than previously published protocols, even higher than those that utilized lentiviral vectors (Levine et al., 2006). This transduction protocol will be clinically relevant with αCD3/αCD28 stimulation and one round of transduction during culture of cells on surfaces coated with Retronectin, in contrast to other protocols that use co-culture with retroviral packaging cell lines to transduce cells in vitro (Matheux et al., 2000; Berger et al., 2001). Those trials using phosphate-depleted media and centrifugation also obtained strong in vitro marking after three rounds of transduction (Bunnell et al., 1997; Donahue et al., 1998; Hanazono et al., 1999; Morgan et al., 2005). However, these were not as high as obtained when using Retronectin and one round of transduction (this study; Levine et al., 2006). In addition to the increased transduction of CD4+ T cells provided by the Retronectin, preloading of the viral particles also concentrated the viral particles from a low-titer producer cell line and removed potential toxic metabolites from the culture supernatant. However, multiple factors besides the frequency of gene transfer into the CD4+ T cells could contribute to the levels of long-term gene marking in vivo. The purified CD4+ T cells were treated with anti-CD3 and anti-CD28 antibodies to simulate natural activation with the addition of IL-2. To function in vivo, these activated and transduced CD4+ T cells are required to return to quiescent state and distribute normally within the blood and secondary lymphoid tissues. We also lowered the IL-2 concentration to 20 U/ml for the remaining 3 days of in vitro culture, but do not have any data on how this contributed to the engraftment of transduced cells. In some cases, nonmyeloablative preconditioning of the host with chemotherapy may increase engraftment of transduced cells (Berger et al., 2001). Because the effects on engraftment are difficult to quantify in an autologous environment, additional studies will be required to optimize long-term gene marking in large animal models.
A number of HIV gene therapy strategies have moved from in vitro experiments into large animal models or clinical trials with limited success—even using inhibitors with strong in vitro effects (Woffendin et al., 1996; Matheux et al., 2000; Ngok et al., 2004; Morgan et al., 2005; Macpherson et al., 2005; Levine et al., 2006; van Lunzen et al., 2007; DiGiusto et al., 2010). Early studies may have been limited by insufficient gene marking or engraftment of manipulated cells (Donahue et al., 1996, 1998; Woffendin et al., 1996; Matheux et al., 2000; Ngok et al., 2004; Macpherson et al., 2005; Morgan et al., 2005; van Lunzen et al., 2007; Mitsuyasu et al., 2009; DiGiusto et al., 2010). In our studies, we found that a single infusion of transduced CD4+ T cells led to high levels of gene marking in vivo, superior to many studies (Woffendin et al., 1996; Matheux et al., 2000; Macpherson et al., 2005; Morgan et al., 2005; van Lunzen et al., 2007) and at least compared to other published studies using retroviral vectors (Bunnell et al., 1997; Donahue et al., 1998; Levine et al., 2006). In those studies, a single infusion of transduced cells led to low but detectable marking, but multiple infusions of transduced cells achieved comparable levels of gene marking (Bunnell et al., 1997). Only with the use of lentiviral vectors in a single infusion of transduced cells were higher levels of in vivo gene marking achieved (Levine et al., 2006). The initial levels of gene marking in PBMCs declined slightly over the first week and were readily detectable in secondary lymphoid tissues. We could not discern whether this decline was a further dissemination of the transduced cells to peripheral tissues or a loss of transduced cells to apoptosis. Long-term analysis of the in vivo gene marking in these studies is complicated by the rapid decline in the CD4+ T-cell populations caused by SHIV viral challenge, a decline that is more rapid than with SIV challenge used in similar studies (Donahue et al., 1998). Interestingly, we observed a transient rise in gene marking approximately 6 weeks post challenge. One possible explanation for this observation is the homeostatic expansion of the lymphocyte populations after the massive decline. The increase in gene marking also suggests functional expansion of the transduced cells in vivo, although we cannot formally exclude redistribution of transduced cells as the cause. These high levels of gene marking in vitro and in vivo achieved with our transduction protocol may be useful for gene therapy of other disease indications.
Given the low levels of gene transfer achieved by most protocols in vivo, selection of transduced cells during viral replication has been proposed as an important component of AIDS gene therapy strategy (Braun and Johnson 2006; Kimpel et al., 2010). Selection of transduced cells has been predicted to be stronger when the inhibitor prevents viral infection rather than just interferes with viral production (von Laer et al., 2006). This would be correct if death of CD4+ T lymphocytes in HIV infection was primarily due to direct infection; however, some reports suggest that toxicity to uninfected bystander CD4+ T cells may also play an important role in the depletion of CD4+ T cells (Clerici et al., 2003). Only a few anti-HIV gene therapy clinical trials (Woffendin et al., 1996; Morgan et al., 2005; Podsakoff et al., 2005) have shown a selective advantage of the therapeutic vector in vivo, whereas no clear evidence for a preferential survival of cells expressing anti-HIV genes was observed in a number of other trials (Donahue et al., 1998; Kang et al., 2002; Amado et al., 2004; Levine et al., 2006; van Lunzen et al., 2007; Mitsuyasu et al., 2009; DiGiusto et al., 2010). As with the clinical trials, most studies in nonhuman primates have not found selective expansion of transduced cells in vivo (Bunnell et al., 1997; Donahue et al., 1998). Selection of a mutant MGMT gene and the envelope-fusion inhibitor increase vector frequency after chemotherapy and SHIV challenge in the nonhuman primate model (Trobridge et al., 2009), as we have also observed with the envelope-fusion inhibitor in vitro (Kimpel et al., 2010) and in these studies with the huTat2 intrabody in vivo. Other strategies, such as those that target muscle cells with adeno-associated viral vectors expressing neutralizing antibodies, may not require selection of transduced cells (Johnson et al., 2009; Balazs et al., 2012). Although a selective survival of huTat2-transduced cells was observed in both animals, detectable expression of the secreted intrabodies was only found in the plasma of the one animal—the animal with lower levels of initial gene marking but higher levels of in vitro expression. As in vivo gene marking improves, we may begin to assess which viral inhibitors effectively protect cells from viral toxicity, provide for the expansion of transduced cells, and will eventually provide a therapeutic benefit.
In summary, while anti-retroviral therapy has been successful in lowering viral loads and extending the life of HIV patients, current therapies require life-long action and complex drug regimens to prevent the disease from progressing to more advanced stages of immunodeficiency. Effective antiretroviral therapy is hindered by the emergence of drug-resistant viral variants and by residual latently infected reservoir populations (Kozal, 2009). Therefore, alternative approaches of therapy such as gene therapy need to be explored. The current study demonstrates in the rhesus macaque animal model that retrovirus-mediated delivery of the anti-Tat intrabody huTat2 is efficient and provides a relative survival advantage for huTat2 intrabody-transduced cells. However, given the discordance between gene marking and viral load, we cannot conclude the huTat2 intrabody is solely responsible for the stabilized CD4+ T cell counts and the control of SHIV replication in this animal. Higher levels of expression may be required to counteract the viral replication that occurs in nontransduced cells. These studies bring to light the difficulty in achieving therapeutic levels of gene marking and expression, and also of translating in vitro inhibition into clinical success. It may be that inhibitors that block multiple causes of HIV pathogenicity, potentially in combination with other modes of viral inhibition, will be necessary in future gene therapy studies. The hope is that one of these gene therapy approaches will complement other treatment modalities and translate into an effective and life-long clinical benefit for patients with HIV disease.
Acknowledgments
These studies were supported by National Institutes of Health (NIH) grants RR14447, AI 61797, CA 73473, RR00164, and RR00168, and by a developmental award from the Partners/Fenway/Shattuck Center for AIDS Research (CFAR), an NIH-funded program (AI 42851). We thank Johnson Wong (Massachusetts General Hospital) for the 6G12 anti-CD3 hybridoma, Dr. M. Gately (Hoffman-La Roche) for the IL-2, Keith Reimann (Beth Israel Deaconess Medical Center, Harvard Medical School) for the SHIV 89.6p challenge stock, Richard C. Mulligan (Harvard Medical School) for the U2OS human osteosarcoma cell line, David A. Williams (Children's Hospital Boston) for the HEL-GFP cell line, and Garry Nolan (Stanford University School of Medicine) for the LZRS vector and the Phoenix packaging cell line. We also thank Lynda Fernsten and the NEPRC Primate Medicine staff for expert care of the animals, Jackie Gillis and Michelle Connole for assistance with flow cytometry.
Author Disclosure Statement
No competing financial interests exist.
References
- Amado R.G. Mitsuyasu R.T. Rosenblatt J.D., et al. Anti-human immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a phase I study: myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Hum. Gene Ther. 2004;15:251–262. doi: 10.1089/104303404322886101. [DOI] [PubMed] [Google Scholar]
- Apolloni A. Hooker C.W. Mak J. Harrich D. Human immunodeficiency virus type 1 protease regulation of tat activity is essential for efficient reverse transcription and replication. J. Virol. 2003;77:9912–9921. doi: 10.1128/JVI.77.18.9912-9921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahner I. Sumiyoshi T. Kagoda M., et al. Lentiviral vector transduction of a dominant-negative Rev gene into human CD34+ hematopoietic progenitor cells potently inhibits human immunodeficiency virus-1 replication. Mol. Ther. 2007;15:76–85. doi: 10.1038/sj.mt.6300025. [DOI] [PubMed] [Google Scholar]
- Bai J. Sui J. Zhu R.Y., et al. Inhibition of Tat-mediated transactivation and HIV-1 replication by human anti-hCyclinT1 intrabodies. J. Biol. Chem. 2003;278:1433–1442. doi: 10.1074/jbc.M208297200. [DOI] [PubMed] [Google Scholar]
- Balazs A.B. Chen J. Hong C.M., et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. Gene therapy. Intracellular immunization. Nature. 1988;335:395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- Berger C. Huang M.L. Gough M., et al. Nonmyeloablative immunosuppressive regimen prolongs In vivo persistence of gene-modified autologous T cells in a nonhuman primate model. J. Virol. 2001;75:799–808. doi: 10.1128/JVI.75.2.799-808.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L. Dykxhoorn D.M. Benita Y., et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Braun S.E. Johnson R.P. Setting the stage for bench-to-bedside movement of anti-HIV RNA inhibitors-gene therapy for AIDS in macaques. Front. Biosci. 2006;11:838–851. doi: 10.2741/1841. [DOI] [PubMed] [Google Scholar]
- Bunnell B.A. Metzger M. Byrne E., et al. Efficient in vivo marking of primary CD4+ T lymphocytes in nonhuman primates using a gibbon ape leukemia virus-derived retroviral vector. Blood. 1997;89:1987–1995. [PubMed] [Google Scholar]
- Clerici M. Villa M.L. Trabattoni D. Shearer G.M. The breakdown of the cytokine network subsequent to human immunodeficiency virus infection. J. Gen. Virol. 2003;64:1649–1661. doi: 10.1155/S0962935195000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiusto D.L. Krishnan A. Li L., et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000931. 36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue R.E. Kirby M.R. Metzger M.E., et al. Peripheral blood CD34+ cells differ from bone marrow CD34+ cells in Thy-1 expression and cell cycle status in nonhuman primates mobilized or not mobilized with granulocyte colony-stimulating factor and/or stem cell factor. Blood. 1996;87:1644–1653. [PubMed] [Google Scholar]
- Donahue R.E. Bunnell B.A. Zink M.C., et al. Reduction in Siv replication in rhesus macaques infused with autologous lymphocytes engineered with antiviral genes. Nat. Med. 1998;4:181–186. doi: 10.1038/nm0298-181. [DOI] [PubMed] [Google Scholar]
- Galati D. Bocchino M. Paiardini M., et al. Cell cycle dysregulation during HIV infection: perspectives of a target based therapy. Curr Drug Targets Immune Endocr. Metab. Disord. 2002;2:53–61. [PubMed] [Google Scholar]
- Gennari F. Mehta M. Wang Y., et al. Direct phage to intrabody screening (DPIS): demonstration by isolation of cytosolic intrabodies against the TES1 site of Epstein Barr virus latent membrane protein 1 (LMP1) that block NF-κB transactivation. J. Mol. Biol. 2004;335:193–207. doi: 10.1016/j.jmb.2003.09.073. [DOI] [PubMed] [Google Scholar]
- Gerard C.J. Arboleda M.J. Solar G., et al. A rapid and quantitative assay to estimate gene transfer into retrovirally transduced hematopoietic stem/progenitor cells using a 96-well format PCR and fluorescent detection system universal for MMLV-based proviruses. Hum. Gene Ther. 1996;7:343–354. doi: 10.1089/hum.1996.7.3-343. [DOI] [PubMed] [Google Scholar]
- Hanazono Y. Brown K.E. Handa A., et al. In vivo marking of rhesus monkey lymphocytes by adeno-associated viral vectors: direct comparison with retroviral vectors. Blood. 1999;94:2263–2270. [PubMed] [Google Scholar]
- Hanenberg H. Hashino K. Konishi H., et al. Optimization of fibronectin-assisted retroviral gene transfer into human CD34+ hematopoietic cells. Hum. Gene Ther. 1997;8:2193–2206. doi: 10.1089/hum.1997.8.18-2193. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Harrich D. McMillan N. Munoz L., et al. Will diverse Tat interactions lead to novel aniretroviral drug targets? Curr. Drug Targets. 2006;7:1595–1606. doi: 10.2174/138945006779025338. [DOI] [PubMed] [Google Scholar]
- Humeau L.M. Binder G.K. Lu X., et al. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol. Ther. 2004;9:902–913. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Jacque J.M. Triques K. Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.R. Schnepp B.C. Zhang J., et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against Siv infection in monkeys. Nat. Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E.M. De Witte M. Malech H., et al. Nonmyeloablative conditioning followed by transplantation of genetically modified HLA-matched peripheral blood progenitor cells for hematologic malignancies in patients with acquired immunodeficiency syndrome. Blood. 2002;99:698–701. doi: 10.1182/blood.v99.2.698. [DOI] [PubMed] [Google Scholar]
- Kawai T. Wong J. Maclean J., et al. Characterization of a monoclonal antibody (6G12) recognizing the cynomolgus monkey CD3 antigen. Transplant. Proc. 1994;26:1845–1846. [PubMed] [Google Scholar]
- Kimpel J. Braun S.E. Qiu G., et al. Survival of the fittest: positive selection of CD4+ T cells expressing a membrane-bound fusion inhibitor following HIV-1 infection. PLoS One. 2010;5:e12357. doi: 10.1371/journal.pone.0012357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella T.M. Nolan G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Klebba C. Ottmann O.G. Scherr M., et al. Retrovirally expressed anti-HIV ribozymes confer a selective survival advantage on CD4+ T cells in vitro. Gene Ther. 2000;7:408–416. doi: 10.1038/sj.gt.3301094. [DOI] [PubMed] [Google Scholar]
- König R. Zhou Y. Elleder D., et al. Global Analysis of Host-Pathogen Interactions that Regulate Early-Stage HIV-1 Replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozal MJ. Drug-resistant human immunodefiency virus. Clin. Microbiol. Infect. 2009;15(Suppl 1):69–73. doi: 10.1111/j.1469-0691.2008.02687.x. [DOI] [PubMed] [Google Scholar]
- Kumar P. Ban H.S. Kim S.S., et al. T cell-specific sirna delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B.L. Humeau L.M. Boyer J., et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lo A. Zhu Q. Marasco W.A. Intracellular antibodies (intrabodies) and their therapeutic potential. Handb. Exp. Pharmacol. 2008;181:343–373. doi: 10.1007/978-3-540-73259-4_15. [DOI] [PubMed] [Google Scholar]
- Macpherson J.L. Boyd M.P. Arndt A.J., et al. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J. Gene Med. 2005;7:552–564. doi: 10.1002/jgm.705. [DOI] [PubMed] [Google Scholar]
- Marasco W.A. Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 2007;25:1421–1434. doi: 10.1038/nbt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco W.A. Lavecchio J. Winkler A. Human anti–HIV-1 tat sFv intrabodies for gene therapy of advanced HIV-1-infection and AIDS. J. Immunol. Methods. 1999;231:223–238. doi: 10.1016/s0022-1759(99)00159-3. [DOI] [PubMed] [Google Scholar]
- Matheux F. Lauret E. Rousseau V., et al. Simian immunodeficiency virus resistance of macaques infused with interferon beta-engineered lymphocytes. J. Gen. Virol. 2000;81:2741–2750. doi: 10.1099/0022-1317-81-11-2741. [DOI] [PubMed] [Google Scholar]
- Mhashilkar A.M. Bagley J. Chen S.Y., et al. Inhibition of HIV-1 Tat-mediated Ltr transactivation and HIV-1 infection by anti-Tat single chain intrabodies. EMBO J. 1995;14:1542–1551. doi: 10.1002/j.1460-2075.1995.tb07140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhashilkar A.M. Lavecchio J. Eberhardt B., et al. Inhibition of human immunodeficiency virus type 1 replication in vitro in acutely and persistently infected human CD4+ mononuclear cells expressing murine and humanized anti-human immunodeficiency virus type 1 Tat single-chain variable fragment intrabody. Hum. Gene Ther. 1999;10:1453–1467. doi: 10.1089/10430349950017798. [DOI] [PubMed] [Google Scholar]
- Michienzi A. Castanotto D. Lee N., et al. RNA-mediated inhibition of HIV in a gene therapy setting. Ann. N. Y. Acad. Sci. 2003;1002:63–71. doi: 10.1196/annals.1281.008. [DOI] [PubMed] [Google Scholar]
- Mitsuyasu R.T. Merigan T. Carr A., et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R.A. Walker R. Carter C.S., et al. Preferential survival of CD4+ T lymphocytes engineered with anti-human immunodeficiency virus (HIV) genes in HIV-infected individuals. Hum. Gene Ther. 2005;16:1065–1074. doi: 10.1089/hum.2005.16.1065. [DOI] [PubMed] [Google Scholar]
- Nathans R. Cao H. Sharova N., et al. Small-molecule inhibition of HIV-1 Vif. Nat. Biotechnol. 2008;26:1187–1192. doi: 10.1038/nbt.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngok F.K. Mitsuyasu R.T. Macpherson J.L., et al. Clinical gene therapy research utilizing ribozymes: application to the treatment of HIV/AIDS. Methods Mol Biol. 2004;252:581–598. doi: 10.1385/1-59259-746-7:581. [DOI] [PubMed] [Google Scholar]
- Nishimura Y. Brown C.R. Mattapallil J.J., et al. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8000–8005. doi: 10.1073/pnas.0503233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina C.D. Murray M.F. Dykxhoorn D.M., et al. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Palmer E. Liu H. Khan F., et al. Enhanced cell-free protein expression by fusion with immunoglobulin Cκ domain. Protein Sci. 2006;15:2842–2846. doi: 10.1110/ps.062429906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi F. The multiple functions of HIV-1 Tat: proliferation versus apoptosis. Front. Biosci. 2006;11:708–717. doi: 10.2741/1829. [DOI] [PubMed] [Google Scholar]
- Pitcher C.J. Hagen S.I. Walker J.M., et al. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- Podsakoff G. Engel B. Carbonaro D.A., et al. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34(+) cells. Mol. Ther. 2005;12:77–86. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Pollok K.E. Hanenberg H. Noblitt T.W., et al. High-efficiency gene transfer into normal and adenosine deaminase-deficient T lymphocytes is mediated by transduction on recombinant fibronectin fragments. J. Virol. 1998;72:4882–4892. doi: 10.1128/jvi.72.6.4882-4892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznansky M.C. Foxall R. Mhashilkar A., et al. Inhibition of human immunodeficiency virus replication and growth advantage of CD4+ T cells from HIV-infected individuals that express intracellular antibodies against HIV-1 gp120 or Tat. Hum. Gene Ther. 1998;9:487–496. doi: 10.1089/hum.1998.9.4-487. [DOI] [PubMed] [Google Scholar]
- Poznansky M.C. La Vecchio J. Silva-Arietta S., et al. Inhibition of human immunodeficiency virus replication and growth advantage of CD4+ T cells and monocytes derived from CD34+ cells transduced with an intracellular antibody directed against human immunodeficiency virus type 1 Tat. Hum. Gene Ther. 1999;10:2505–2514. doi: 10.1089/10430349950016843. [DOI] [PubMed] [Google Scholar]
- Romani B. Engelbrecht S. Glashoff R.H. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J. Gen. Virol. 2010;91:1–12. doi: 10.1099/vir.0.016303-0. [DOI] [PubMed] [Google Scholar]
- Rossi J.J. June C.H. Kohn D.B. Genetic therapies against HIV. Nat. Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver N. Cantin E.M. Chang P.S., et al. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- Sodora D.L. Lee F. Dailey P.J. Marx P.A. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res. Hum. Retroviruses. 1998;14:171–181. doi: 10.1089/aid.1998.14.171. [DOI] [PubMed] [Google Scholar]
- Swan C.H. Bühler B. Steinberger P., et al. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther. 2006;13:1480–1492. doi: 10.1038/sj.gt.3302801. [DOI] [PubMed] [Google Scholar]
- The Institute Of Laboratory Animal Research COLS, National Research Council. Guide for the Care and Use of Laboratory Animals. National Academy Press; Washington, DC: 1996. [Google Scholar]
- Trobridge G.D. Wu R.A. Beard B.C., et al. Protection of stem cell-derived lymphocytes in a primate AIDS gene therapy model after in vivo selection. PLoS One. 2009;4:e7693. doi: 10.1371/journal.pone.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lunzen J. Glaunsinger T. Stahmer I., et al. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol. Ther. 2007;15:1024–1033. doi: 10.1038/mt.sj.6300124. [DOI] [PubMed] [Google Scholar]
- Veazey R.S. Tham I.C. Mansfield K.G., et al. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early Siv infection in vivo. J. Virol. 2000;74:57–64. doi: 10.1128/jvi.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Laer D. Hasselmann S. Hasselmann K. Impact of gene-modified T cells on HIV infection dynamics. J. Theor. Biol. 2006;238:60–77. doi: 10.1016/j.jtbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Woffendin C. Ranga U. Yang Z., et al. Expression of a protective gene-prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. Murakami A. Johnson R.P., et al. Optimization of ex vivo activation and expansion of macaque primary CD4-enriched peripheral blood mononuclear cells for use in anti-HIV immunotherapy and gene therapy strategies. J. Acquir. Immune Defic. Syndr. 2003;32:245–254. doi: 10.1097/00126334-200303010-00002. [DOI] [PubMed] [Google Scholar]