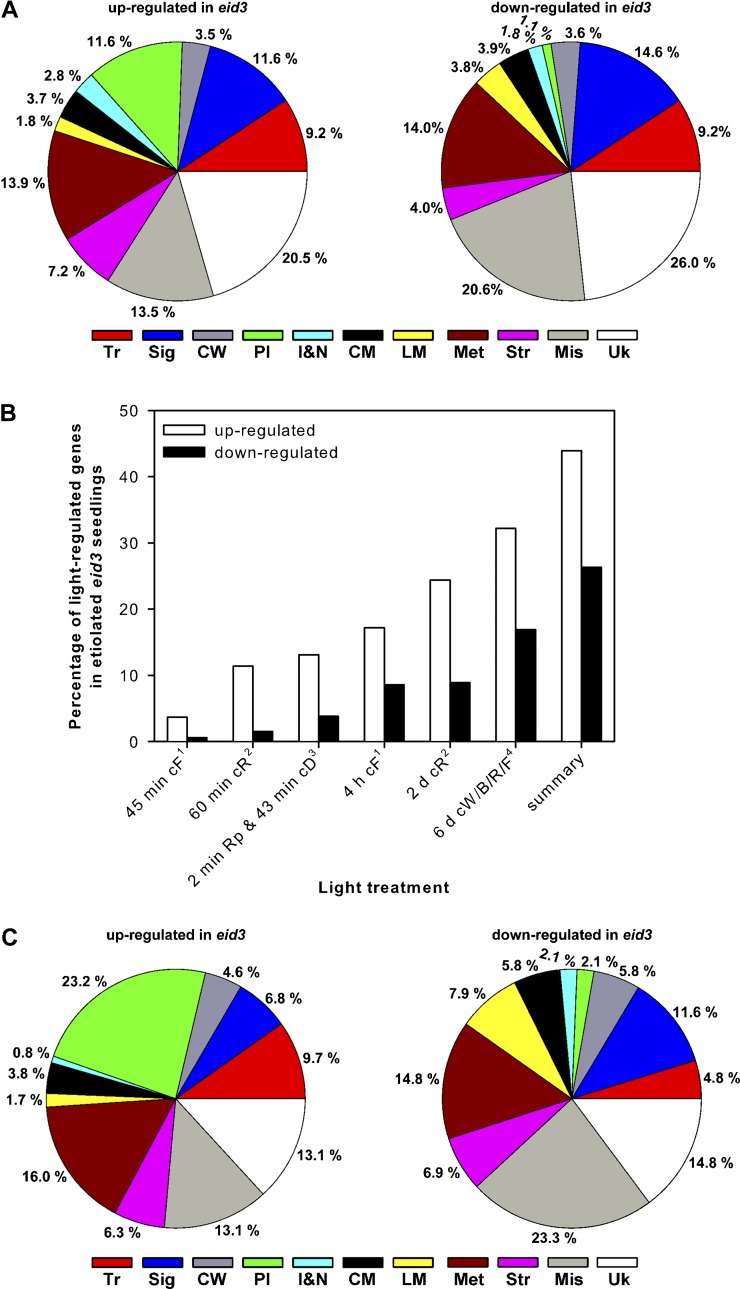
Figure 6.
Analyses of transcript accumulation patterns in dark-grown eid3 seedlings. A, Ler wild-type and eid3 seeds were grown in darkness for 4 d before extraction of RNA and determination of transcript accumulation patterns by microarray analyses. Pie charts show the distribution of more than 2-fold up- or down-regulated genes among functional categories. Distributions are expressed as percentages of 541 up-regulated and 718 down-regulated genes in etiolated eid3 compared with wild-type seedlings. B, Bar charts depict percentages of up- and down-regulated genes in etiolated eid3 seedlings that have been annotated as being light regulated in published data sets (Jiao et al., 2005; Leivar et al., 2009; Peschke and Kretsch, 2011) and upon red light pulse treatment of 4-d-old, etiolated Ler wild-type seedlings (this study). Ler wild-type seedlings received one saturating red light pulse before transfer back to darkness for 43 min. Transcript levels were compared with etiolated seedlings harvested at the same time. Genes exhibiting more than 2-fold up- or down-regulation in transcript levels were annotated as being light regulated: 1Peschke and Kretsch (2011); 2Leivar et al. (2009); 3this study; 4Jiao et al. (2005). cD, Continuous darkness; cF, continuous far-red light; cR, continuous red light; cW/B/R/F, continuous white/blue/red/far-red light; Rp, red light pulse. C, Pie charts show the distribution among functional categories of up- and down-regulated genes in etiolated eid3 seedlings that have been annotated as being light regulated in other data sets. Distributions are expressed as percentages of 337 up-regulated and 179 down-regulated genes. Tr, Transcription; Sig, signaling; CW, cell wall; Pl, plastids; I&M, ions and nutrition; CM, carbohydrate metabolism; LM, lipid metabolism; Met, metabolism miscellaneous; Str, stress; Mis, miscellaneous; Uk, unknown.