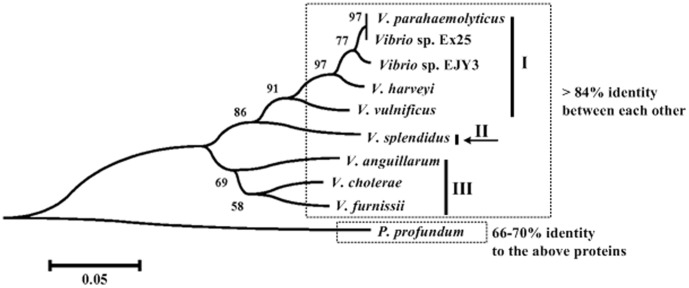
Figure 2. Phylogenetic tree of AphA orthologs.
The protein sequences were derived from V. parahaemolyticus RIMD 2210633 [14], Vibrio sp. Ex25 (Accession numbers CP001805.1, and CP001806.1), Vibrio sp. EJY3 [48], V. harveyi ATCC BAA-1116 [49], V. vulnificus YJ016 [50], V. splendidus LGP32 [51], V. anguillarum 775 [52], V. cholerae N16961 [53], V. furnissii NCTC 11218 [54], and P. profundum SS9 [25]. The a.a. sequences were aligned by the CLUSTALW [55] web server at http://align.genome.jp/. The aligned sequences were then used to construct an unrooted neighbor-joining tree using MEGA version 5.0 [56] with a bootstrap iteration number of 1000. Shown on branch points of phylogenic tree were bootstrap values (%).