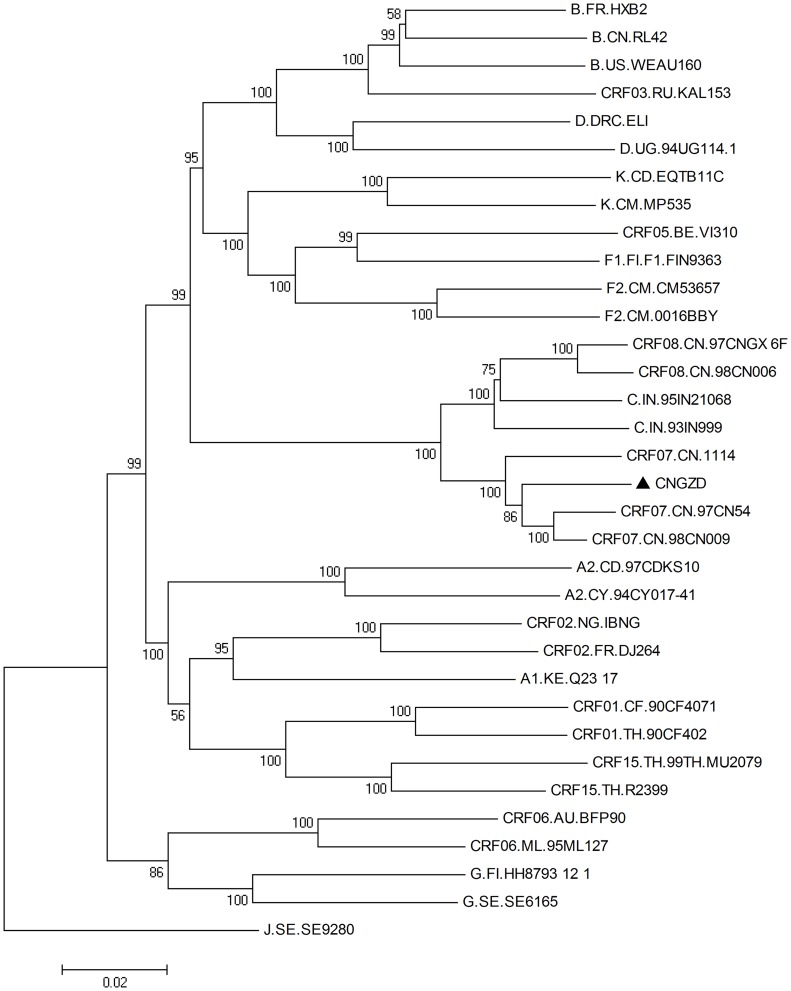
Figure 3. Phylogenetic analysis of clinical isolated virus strain CNGZD.
A neighbor joining phylogenetic tree was built by Mega 4.1 based on full length sequences of clinical isolated virus strain CNGZD and different pure subtypes and CRFs. The Kimura 2-parameter substitution model was used and bootstrap values (1000 replicates) higher than 50 were shown next to the nodes of the tree. J.SE. SE9280 with long distance to CRF07_BC was selected as an outgroup in the phylogentic tree. Subtype references were downloaded from the Los Alamos HIV database (http://hiv-web.lanl.gov). CNGZD located in a clade with the other CRF07_BC isolates. Bar: 0.002 substitutions per site.