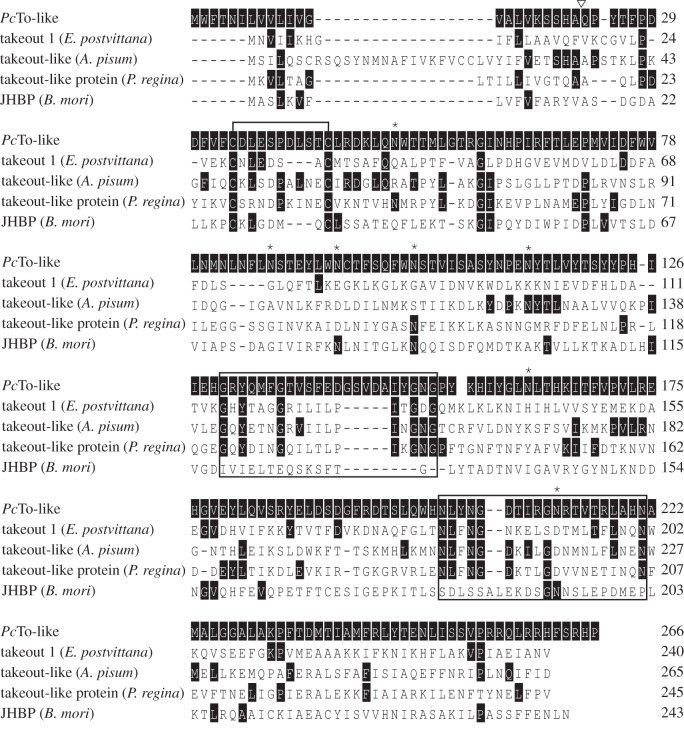
Figure 4.
Amino acid alignments (ClustalW) of PcTo-like from P. cochleariae and other members from the To/JHBP family (Epiphyas postvittana To1: GeneBank: ACF39401; Acyrthosiphon pisum To-like: GeneBank: XP_001952685; Phormia regina To-like protein: GeneBank: BAD83405; Bombyx mori JHBP: GeneBank: AAF19267). Solid black shading depicts amino acids identical to PcTo-like sequence. White inverted triangles indicate the predicted signal peptide of PcTo-like. Asterisks mark the predicted N-glycosylation sites. Conserved cysteine residues that form disulphide bonds are marked with a bracket above the sequence. The two black boxes show the location of the To-typical motives.