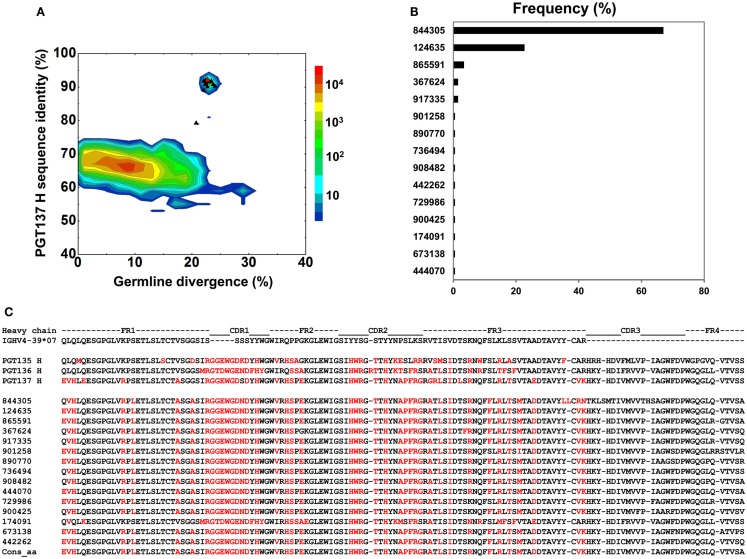
Figure 4.
Sequence selection for functional characterization of heavy chains from donor 39. (A) Divergence/identity analysis of 15 heavy-chain variable-domain sequences obtained from the clustering analysis of 202 sequences identified by intra-donor phylogenetic analysis. Sequences of IGHV4-39 origin are plotted as a function of sequence identity to PGT137 heavy chain and sequence divergence from inferred germline allele, with 15 selected sequences shown as black triangles and their amino-acid consensus as red triangle. (B) Percent population of 15 clusters obtained using a sequence identity cutoff of 95%. Each cluster is indicated by its representative sequence. “Frequency” refers to the total number of sequences observed for each cluster. (C) Protein sequences of 15 cluster representatives and their amino-acid consensus. Sequences are aligned to the inferred germline gene, IGHV4-39*07. Framework regions (FR) and complementarity-determining regions (CDRs) are based on Kabat nomenclature. Amino acids mutated from the germline gene are shown in red.