Figure 6. Genes predicted to be essential for Mycobacterium tuberculosis H37Rv to adapt to hypoxia.
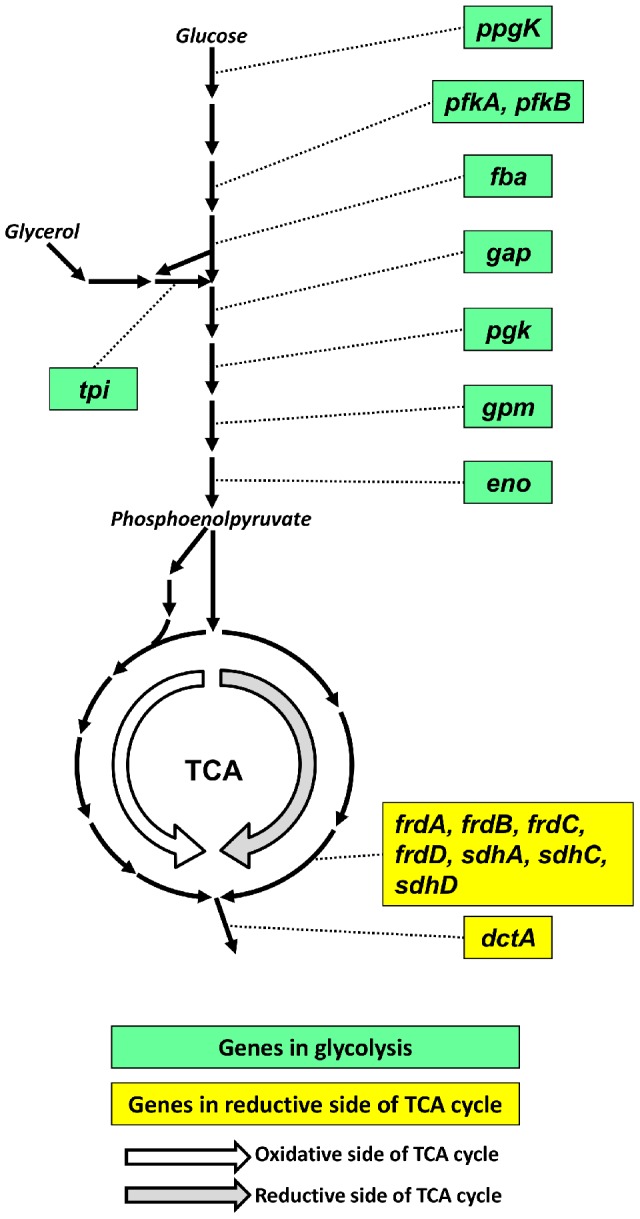
Shown are the genes predicted to be nonessential under normoxia but essential under hypoxia for the wild type strain. Given the metabolic state shown in Figure 5, the genes predicted to be essential for hypoxic adaptation were mostly located in the glucose/glycerol processing pathways and on the reductive side of the tricarboxylic acid (TCA) cycle. dctA, Na+/H+-dicarboxylate symporter; eno, enolase; fba, fructose-bisphosphate aldolase; frdA, frdB, frdC, frdD, fumarate reductase; gap, glyceraldehyde-3-phosphate dehydrogenase; gpm, phosphoglycerate mutase; pfKA, pfkB, phosphofructokinase; pgk, phosphoglycerate kinase; ppgK, polyphosphate glucokinase; sdhA, sdhC, sdhD, succinate dehydrogenase; tpi, triosephosphate isomerase.
