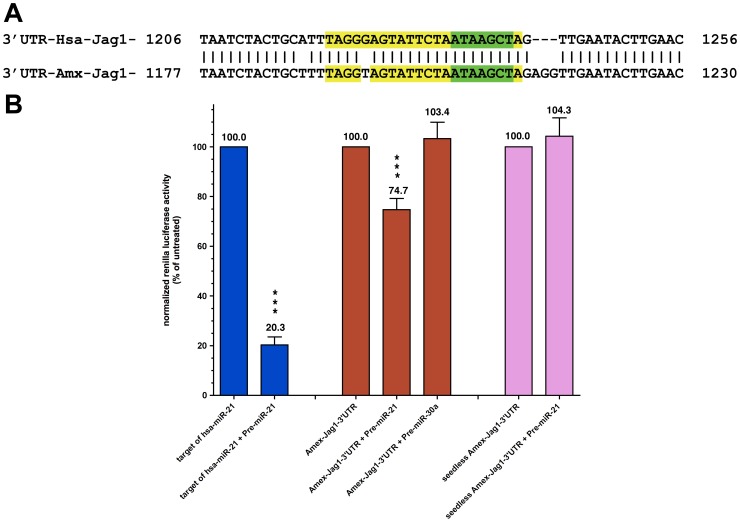
Figure 4. Jagged1 as a putative target of Amex-miR-21.
A. Comparison between the human and axolotl miR-21 target sites in their Jagged1 genes. Nucleotide alignment of the miR-21 target site-containing region present in the 3′-UTR of the human Jagged1 (Hsa-Jag1, NM_000214) and the axolotl Jagged1 (Amx-Jag1, JF907581). The 22 bases comprising the target site for miR-21 are in yellow except for the 7 bases complementary to the miR-21 seed region (green color). Vertical bars (|) denote nucleotide identities between the two sequences. The numbers at both sides of the alignment are the nucleotide position on each 3′-UTR. B. In vitro luciferase assays testing the effect of miR-21 on the 3′-UTR of Amex-Jag1 in axolotl cells. Results are expressed as percent of co-reporter-normalized Renilla activity against reference vectors. Bars denote standard error of mean of the amount of independent assays. Student t-test was done and the obtained p-values determined that the Target of Hsa-miR-21 is a good positive control as biosensor for the activity of this microRNA (p≤0.005; two-tailed t-test). These results suggest that Amex-Jag1 may in fact be a target for miR-21 because significant differences were found in the Renilla signal recorded from cells electroporated with only the vector containing the 3′-UTR of Amex-Jag1, versus the cells that were also electroporated with Pre-miR-21 *** (p≤0.005) but not Pre-miR30a. When the latter experiment was repeated with the mutant, seedless-Amex-Jag1-3′-UTR, any sensitivity to exogenous miR-21 was lost.