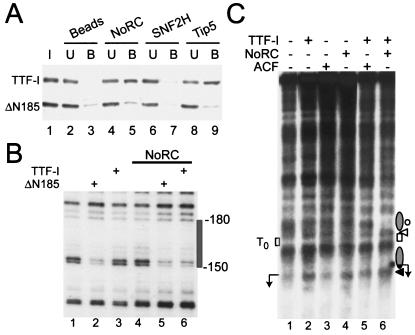
FIG. 2.
NoRC-TTF-I interaction triggers chromatin remodeling at the rDNA promoter. (A) Pull-down assay. NoRC, Snf2H, and Tip5 were immobilized on beads and incubated with a mixture of full-length TTF-I and N-terminally truncated TTF-I (ΔN185). TTF-I in the input (I), unbound (U), and bound to the beads (B) was analyzed by Western blotting. (B) Chromatin was incubated with TTFΔN185 (ΔN185, lanes 2 and 5) and TTF-I (lanes 3 and 6) in the absence or presence of NoRC. TTF binding to T0 was detected by partial MNase digestion of the DNA and primer extension footprinting. The gray box indicates the TTF binding site. (C) Mapping of MNase cleavage sites by indirect end labeling of the rDNA promoter reconstituted into chromatin. Chromatin, in the presence of ATP, was incubated with TTF-I, ACF, or NoRC (lanes 2 to 4) and TTF-I in the presence of ACF or NoRC (lanes 5 and 6). Each reaction was digested with 1 U of MNase for 30 s, and purified DNA was digested with NdeI and analyzed by Southern blotting. The positions of rearranged nucleosomes (gray ellipse), the TTF-I binding site, and the transcription start site (arrow) are indicated. Protected (circles) or enhanced (white and black triangles) MNase cleavage sites are indicated on the right. Position +22, the 3′ boundary of the nucleosome positioned at the rDNA promoter, is marked by the black triangle.