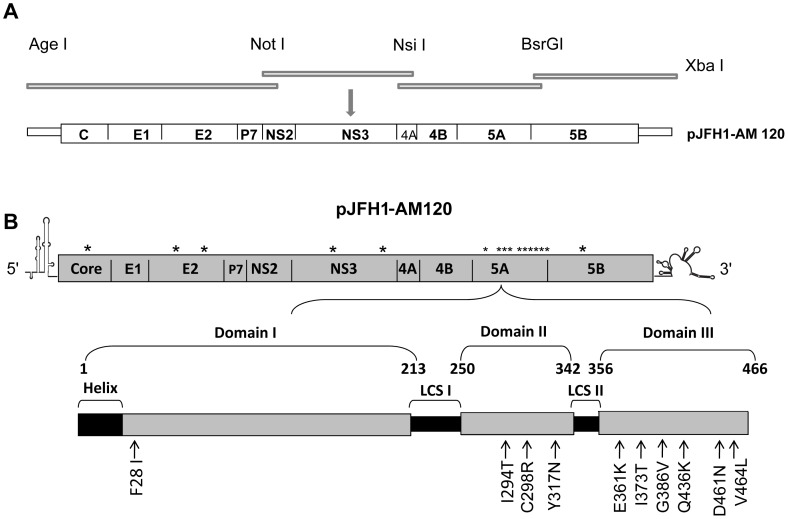
Figure 2. Sequence analysis of the adapted HCV JFH1 variant (JFH1-AM120) and construction of pJFH1-AM120.
(Panel A).Total RNA was isolated from confluent Huh-7.5 cells infected with the 120th day adapted virus (Methods). The total RNA was reverse transcribed with random primers and the resulting cDNA was used as a template for subsequent PCR amplification of four overlapping amplicons (Panel A) (Methods). Four fragments of HCV cDNA (nt1–3059, 2880–5400, 5233–7816 and 7645–9678) were amplified to cover the entire HCV genome. The PCR amplicons were sequenced and the four fragments were sequentially sub-cloned into pJFH1 using the unique restriction enzyme sites Age I/Not I, Not I/Nsi I, Nsi I/BsrG I and BsrG I/Xba I. The plasmid encoding the adapted pJFH1 variant was designated JFH1-AM120 (Panel B). Location of adaptive mutations in JFH1-AM120 are indicated by *.