Abstract
Dendritic cell (DC) derived cytokines play a key role in specifying adaptive immune responses tailored to the type of pathogen encountered and the local tissue environment. However, little is known about how DC perceive the local environment. We investigated whether endogenous Notch signaling could affect DC responses to pathogenic stimuli. We demonstrate that concurrent Notch and TLR stimulation results in a unique cytokine profile in mouse bone-marrow derived DC characterized by enhanced IL-10 and IL-2 and reduced IL-12 expression compared to TLR ligation alone. Unexpectedly, modulation of cytokine production occurred through a non-canonical Notch signaling pathway, independent of γ-secretase activity. Modulation required de novo protein synthesis and PI3K, JNK and ERK activity were necessary for enhanced IL-2 expression while modulation of IL-10 only required PI3K activity. Further, we show that this γ-secretase independent Notch pathway can induce PI3K activity. In contrast, expression of the canonical Notch target gene Hes1 was suppressed in DC stimulated with Notch and TLR ligands simultaneously. Thus, our data suggest that Notch acts as an endogenous signal that modulates cytokine expression of DC through a non-canonical pathway and therefore has the potential to tailor the subsequent adaptive immune response in a tissue and/or stage dependent manner.
Introduction
Dendritic cells (DC3) act to bridge the innate and adaptive immune responses. In their immature state, DC are highly specialized for antigen uptake and detection of pathogen associated molecular patterns through the expression of a wide range of receptors on their surface, such as the Toll-like receptor family (TLR), and form a surveillance network across virtually all tissues (1). Upon detection of a pathogenic stimulus DC are transformed into potent inducers of naïve T cell activation and differentiation, and further influence T cell fate via the production of polarizing cytokines (2, 3). The cytokine signature of mature DC depends on the type of pathogen perceived and the environment it was perceived in, however, little is known about how DC integrate different types of pathogen and environmental signals and how these signals determine the resulting DC cytokine signature.
Notch signaling during embryogenesis provides both temporal and spatial cues that are critical for embryonic development of all animals (4). Temporal and spatial regulation of Notch activation is achieved through differential expression of Notch ligands, receptors and modulators in a tissue and developmental stage specific manner. In adult organisms Notch signaling continues to play a vital role in regulating differentiation decisions in self-renewing tissues such as the hematopoietic system (5, 6), and Notch components continue to be expressed across the body in a tissue and differentiation stage dependent manner.
Ligation of Notch at the cell surface, by a cell bearing Notch ligands, induces γ-secretase dependent cleavage of Notch that releases the intracellular portion (NIC) from the plasma membrane allowing translocation to the nucleus. Notch signaling directly activates expression of target genes through NIC interaction with the transcriptional switch RBPj, and indirectly mediates repression of other genes through inducing expression of the transcriptional repressor Hes1 (7, 8). In addition to this canonical signaling route, alternative pathways involving RBPj and γ-secretase independent signaling have been reported (9-13). However, relatively little is known about the molecules involved in alternative signaling or their involvement in the immune system.
DC express both Notch ligands and receptors (14, 15) and ligation with recombinant ligands has been shown to induce surface expression of MHC Class II, CD80 and CD86 expression (16, 17). However, Notch ligation did not induce CD40 expression and induced a distinct cytokine profile characterized by IL-2 in the absence of more pro-inflammatory cytokines such as IL-6, IL-12 or IL-23 (16). These Notch conditioned DC could sustain proliferation and suppressive activity of CD25+ regulatory T cells and induced IL-17 expression in these cultures in an IL-2 dependent manner (16). We set out to establish whether Notch signaling could influence DC maturation to pro-inflammatory stimuli. We demonstrate that DC stimulated simultaneously with Notch and TLR ligands have a distinct cytokine profile compared to DC stimulated with either stimulus alone. Modulation of DC responses to TLR ligation occurred via a non-canonical Notch signaling pathway and was dependent on PI3K activity. Further, we demonstrated that non-canonical Notch signaling can increase PI3K activity in DC and thus our data support a model where the Notch and TLR pathways interact in DC to influence the cytokine profile of these cells via convergence on PI3K.
Materials and Methods
Animals
Male C57BL/6 mice (Harlan) were maintained in accordance with UK Home Office guidelines (Animals [Scientific Procedures] Act 1986).
Bone marrow-derived DC (BMDC) cultures
BMDC were prepared from wild-type or IL-10−/− mice (obtained with permission from the MGC Foundation, Germany) as previously described (16).
Recombinant Notch ligand rat Jagged1-humanFc fusion protein (R&D Systems) or Human IgG1 (Sigma) control was immobilized onto tissue culture plates via overnight incubation (10 μg/ml). TLR ligands were added as follows: 100 ng/ml LPS from Escherichia coli O26:B6 (Sigma), 0.5 μg/ml CpG 1826, 1 μg/ml Pam3CSK4, 100 μg/ml Poly [I:C] (Invivogen). BMDC were pre-treated with inhibitors for 30-60 min as specified; 2 μM BAY11-7082, 10 μM LY294002, 5 μM SB203580, 10 μM SP600125, 25 μM U0126, and 10 μM DAPT (Calbiochem), 10 μg/ml Cycloheximide (Sigma).
C. jejuni 11168H was cultured in a variable atmosphere incubator (Don Whitely Scientific, UK) under microaerobic conditions (5% O2, 85% N2, 10% CO2) at 37°C. Initial cultures grown for 24 h on complete blood agar plates supplemented with Skirrow selective supplement (Oxoid, UK) were used to inoculate flasks of MEM-α (Invitrogen) supplemented with 20 mM L-Serine and 40 μM Ferric sulphate to an OD600nm of 0.01 and then grown for a further 16 h shaking at 75 rpm. BMDC were infected at a multiplicity of infection of 100 for 3 h before cells were collected for RNA extraction.
Enzyme-linked immunosorbant assays (ELISA)
ELISA sets were purchased from BD Biosciences.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was prepared as previously described (16). Results were normalized to 18S rRNA content and relative mRNA levels are expressed as fold change compared to the IgG1 control or, when no expression was detected in the IgG1 control, as fold change relative to the limit of detection.
Western Blot
JAWSII DC-like cell line was maintained in RPMI supplemented with 20% FCS, 2mM glutamine, 50 units/ml penicillin and 50 μg/ml streptomycin. Cells were serum starved overnight and pre-treated with DAPT or DMSO (vehicle control) for 30 min at 37°C, and the plated on 6 well plates pre-coated with Jagged1-Fc or IgG1 control with or without LPS. After 30-60 min cells were harvested on ice and lysed in 40 μl cold lysis buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, supplemented with 1x Complete-mini-protease inhibitors (EDTA free, Roche), 0.2 mM sodium orthovanadate, 10 mM sodium fluoride, 2 mM sodium pyrophosphate and 100 nM calyculin A) for 20 min. Protein (25 μg) was loaded onto precast 10% Tris Glycine gels (Invitrogen) and, following electrophoresis, transferred onto nitrocellulose membranes (Hybond ECL, Amersham) and blocked with 5% skimmed milk (Marvel) in TBS with 0.3% Tween-20. Membranes were incubated with anti-phospho Akt antibody (Ser473), anti-pan Akt antibody (both New England Biolabs) or anti-Actin antibody (Millipore) overnight at 4°C. Membranes were washed and then incubated with a HRP conjugated secondary antibody (GE Healthcare) for 2 h. After washing, membranes were incubated with SuperSignal West Pico Chemiluminescence Substrate (Pierce) for 5 min and chemiluminescence was visualized on a ChemiDoc XRS+ system (BioRad).
Statistics
All analyses were carried out using GraphPad Prism® (GraphPad Software Inc). A non-parametric one-way ANOVA (Kruskal-Wallis test) was used in Figure 1. For experiments with inhibitor, C. jejuni, or comparing wild-type and knock-out a two-way ANOVA was used instead.
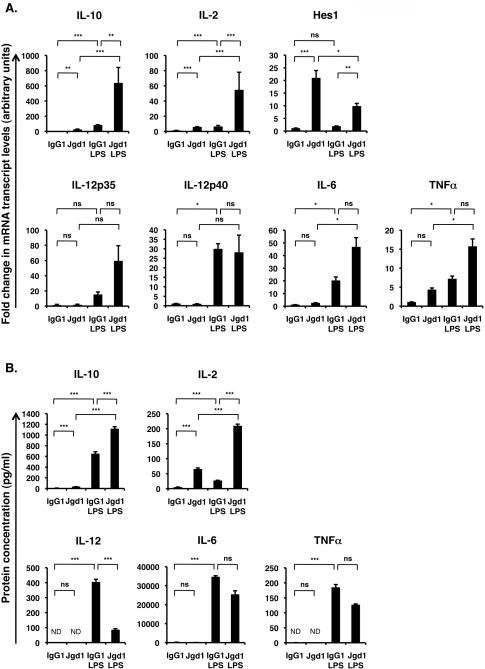
Figure 1. Simultaneous Jagged1-Fc and LPS stimulation of DC preferentially enhances expression of IL-10 and IL-2 while inhibiting IL-12 expression.
BMDC were stimulated with plate bound Jagged1-Fc (Jgd1) or IgG1 (Fc control) in the presence or absence of 100 ng/ml LPS. (A) mRNA transcripts were measured, after 4 h, by qRT-PCR. (B) Accumulation of cytokines in the supernatant, after 24 h, was measured by ELISA. Data are mean ± SD of triplicates and are representative of at least 6 independent experiments. Friedman test with Dunn’s post-test was used to statistically compare each stimulus; P<0.05 was considered significant. * = P<0.05, ** = P<0.01, *** = P<0.001, ns = not significant. ND = not detected.
Results
Notch Signaling modulates DC responses to LPS
To investigate whether Notch signaling could alter the response of DC to pro-inflammatory stimuli, BMDC were stimulated with immobilized recombinant Notch ligand Jagged1-Fc (or whole human IgG1 as an Fc control with similar steric constraints and molecular weight to the Jagged1 fusion protein) with or without LPS. Relative mRNA transcript levels at 4 h and secreted cytokines after 24 h are shown in Figure 1. Hes1, as a known direct target of canonical Notch signaling (18), was measured as a read-out for Notch activation. As expected, Jagged1-Fc induced expression of Hes1 mRNA transcripts (Figure 1B, see also Figure 2 and Supplementary Figure S1). Hey1, Hey 5 and Deltex1 have been reported to be Notch targets in other cell types, however, these were not present at detectable levels in BMDC following stimulation with Jagged1-Fc, LPS or both (data not shown). The relative change in Hes1 mRNA transcripts induced by Jagged1-Fc varied considerably, ranging from 2-30 fold, reflecting the fact that Hes1 transcript and protein expression oscillate with a periodicity of 2 h as a result of Hes1 protein acting as a repressor at its own promoter (19), and thus relative Hes1 mRNA levels will vary with time and degree of synchronization between cells. Addition of the protein synthesis inhibitor cycloheximide (to prevent Hes1 mediated self-repression) to these cultures confirmed that Jagged1-Fc robustly induces Hes1 transcript levels (Supplementary Figure S1A). Furthermore, pre-treatment with the γ-secretase inhibitor DAPT confirmed that expression Hes1 mRNA in response to Jagged1-Fc requires canonical Notch signaling (Supplementary Figure S1B), thus demonstrating that Jagged1-Fc mediates a robust canonical Notch signal.
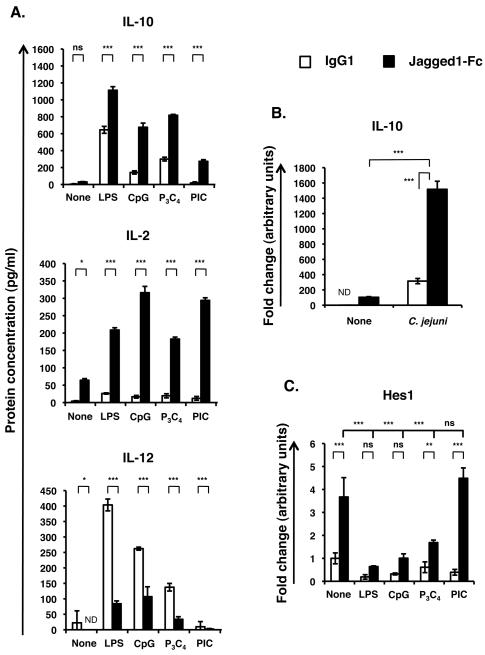
Figure 2. Jagged1-Fc can modulate cytokine expression in response to various TLR ligands and in response to Campylobacter jejuni.
(A) BMDC were stimulated with Jagged1-Fc (black bars) or IgG1 (white bars) in the presence of 100 ng/ml LPS, 0.5 μg/ml CpG, 1 μg/ml Pam3CSK4 (P3C4), 100 μg/ml Poly [I:C] (PIC) or nothing (none) for 24 h and accumulation of IL-10, IL-2 and IL-12 in the supernatant was measured by ELISA. (B) BMDC were stimulated with Jagged1-Fc or IgG1 in the presence or absence of whole, live Campylobacter jejuni at a MOI of 100. (C) BMDC were stimulated as for (A) for 4 h and Hes1 mRNA transcripts were measured by qRT-PCR. Data in all three parts are mean ± SD of triplicates and are representative of at least 3 independent experiments. Two-way ANOVA with Bonferroni post-test was used to statistically compare stimuli; P<0.05 was considered significant. * = P<0.05, ** = P<0.01, *** = P<0.001, ns = not significant. ND = not detected.
Jagged1-Fc alone induced a significant increase in both mRNA and secreted IL-10 and IL-2, but did not significantly induce either mRNA or secreted protein levels of the more pro-inflammatory IL-12, IL-6 or TNFα (Figure 1 and Bugeon et al. (16)). LPS alone increased levels of mRNA transcripts and secreted IL-10, IL-2, IL-12, IL-6 and TNFα compared to control (Figure 1). IL-12p35 transcripts levels were highly variable and were not significantly induced by LPS, which may be consistent with the observation that p35 is mainly regulated at the post-transcriptional level by a series of post-translational modifications (20, 21).
Compared to LPS alone, simultaneous stimulation with Jagged1-Fc and LPS resulted in significantly enhanced expression of IL-10 and IL-2 mRNA and protein, whilst secretion of IL-12 was significantly inhibited (Figure 1). IL-12p35 and p40 mRNA levels were not significantly inhibited by Jagged1-Fc + LPS (compared to LPS alone) suggesting that Jagged1-Fc inhibits LPS induced IL-12 secretion by a post-transcriptional mechanism. IL-6 and TNFα mRNA levels were generally increased by combined Jagged1-Fc and LPS stimulation compared to LPS alone while protein levels were usually reduced, but this was variable and was not statistically significant across all experiments performed. Further, as others have suggested that Hes1 may regulate IL-6 and TNFα mRNA expression (22), we assessed transcript levels in the presence of cycloheximide and found levels unchanged by inhibition of protein translation (Supplementary Figure S1A).
Analysis of Notch receptor transcript levels demonstrated that Notch2 is the predominant Notch receptor present in BMDC, while Notch1 is expressed at only relatively low levels and Notch3 and 4 mRNA transcripts are barely detectable (Supplementary Figure S2A). Notch receptor transcript levels were unaffected by stimulation with TLR ligands (data not shown).
Measurement of mRNA decay rates after stimulation with Jagged1-Fc + LPS or LPS alone for 4 h demonstrated that Jagged1-Fc does not significantly alter the stability of IL-10, IL-2 or IL-12 mRNA (data not shown), suggesting that Jagged1-Fc affects IL-12 expression either at the translational level or through regulation of post-translational modifications, while regulating transcription of IL-10 and IL-2.
In agreement with the results obtained using recombinant Jagged1-Fc as a Notch stimulus, culturing BMDC with a cell-line over expressing the Notch ligand Jagged-1 also enhanced expression of IL-10 in response to LPS (Supplementary Figure S2B). The parental cell line had no effect on LPS induced IL-10 expression, nor did these cells make IL-10 themselves. Thus changes in DC cytokine expression are a result of Notch signaling and not Fc-mediated effects or artifacts due immobilization of Notch ligand to tissue culture plastic.
Additionally, Jagged1-Fc enhanced LPS induced surface expression of CD40 – increasing the percentage of CD40+ DC from 71% to 89% and the mean intensity fluorescence intensity from 87 to 161 (Supplementary Figure S3). Compared to IgG1 control, Jagged1-Fc induced a small increase CD40 expression (from 41% to 58%). However, Jagged1-Fc did not significantly increase CD80, CD86 or MHC Class II surface expression compared to IgG1 stimulated DC, or alter LPS induced changes in these maturation markers (Supplementary Figure S3).
Cross-talk between the Notch and TLR pathways was not limited to modulation of LPS induced cytokine and CD40 expression; combined Jagged1-Fc and LPS stimulation significantly inhibited Hes1 mRNA transcript levels compared to cells stimulated with Jagged1-Fc alone (Figure 1B). Suppression of Hes1 mediated oscillations with cycloheximide confirmed that LPS robustly inhibits induction of this canonical Notch target gene by Jagged1-Fc, by ~4 fold (Supplementary Figure S1A).
Our data suggest Notch and TLR4 signaling in response to Jagged1 and LPS respectively can interact in DC and this interaction is bidirectional – resulting in a modulated cytokine response characterized by enhanced IL-10 and IL-2 but reduced IL-12 expression, as well as reduced expression of the canonical Notch target Hes1.
Notch ligation modulates DC responses to MyD88 dependent and independent TLR agonists and to live C. jejuni
To determine if Notch signaling in response to Jagged1-Fc could modulate the cytokine response of DC to other agonists of the TLRs, BMDC were stimulated with CpG (TLR9 agonist), Pam3CSK4 (TLR2) or Poly [I:C] (TLR3) in the presence of either Jagged1-Fc or IgG1 control. Levels of secreted IL-10, IL-2 and IL-12 are shown in Figure 2A compared to that of BMDC stimulated with LPS (TLR4) or nothing. Similar to LPS stimulated cells, simultaneous stimulation with Jagged1-Fc resulted in significantly enhanced levels of secreted IL-10 and IL-2 in response to CpG, Pam3CSK4 or Poly [I:C]. Poly [I:C] induced relatively little IL-12. In contrast, both CpG and Pam3CSK4 robustly induced IL-12 secretion and this was significantly inhibited by Jagged1-Fc. Thus Notch and TLR signaling can interact in DC through both the MyD88 dependent and the TRIF dependent TLR signaling pathways. This result was confirmed using BMDCs from either MyD88 or TRIF deficient mice (Supplementary Figure S4). Enhanced IL-10 and IL-2 expression in response to combined Notch and TLR2, 4 and 9 stimulation was blocked by MyD88 deficiency, while expression in response to Notch and TLR3 stimulation was unaffected. Conversely TRIF deficiency had no effect on enhanced IL-10 and IL-2 secreted in response to Jagged1-Fc plus CpG or Pam3CSK4, but did prevent enhanced IL-10 and IL-2 in response to combined Notch and TLR3 signaling.
To confirm that Notch signaling can alter responses to whole pathogens, BMDC were stimulated with Jagged1-Fc (or IgG1 control) in the presence or absence of live C. jejuni. DC are thought to be involved in initial responses to C. jejuni, and activation of both TLR2 and 4 have been implicated (23, 24). Notch signaling significantly enhanced IL-10 mRNA transcript levels induced by C. jejuni (Figure 2B).
Jagged1-Fc induced Hes1 expression was significantly inhibited by CpG, Pam3CSK4 and LPS (Figure 2C). In contrast, Poly [I:C] did not inhibit Notch induced Hes1 mRNA transcript levels.
Modulation of the DC cytokine profile by interaction between the Notch and TLR pathways is dependent on de novo protein synthesis, but canonical Notch signaling is dispensable
We next investigated how the Notch and TLR pathways interact at the molecular level to enhance expression of IL-10 and IL-2 whilst suppressing IL-12 expression. To determine whether canonical Notch signaling is required for modulation of DC responses to TLR agonists, BMDC were stimulated with Jagged1-Fc and/or LPS in the presence of the γ-secretase inhibitor DAPT (Figure 3A). DAPT had no effect on expression of either IL-10 or IL-2, indicating that modulation of LPS induced IL-10 and IL-2 by Notch signaling occurs through a non-canonical signaling route. While inhibition of γ-secretase activity caused a significant increase in LPS induced IL-12 secretion, Jagged1-Fc mediated suppression of IL-12 secretion was not abrogated, suggesting that the Notch and TLR pathways interact to modulate the DC cytokine profile through a non-canonical γ-secretase independent route.
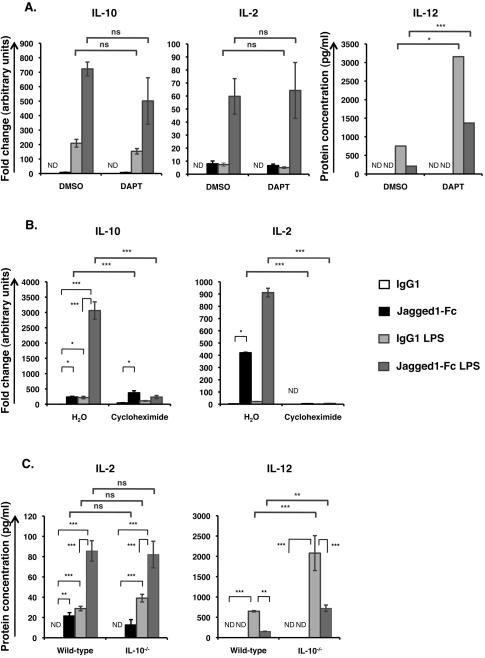
Figure 3. Modulation of TLR induced cytokine expression depends on de novo protein synthesis but is independent of canonical, γ-secretase mediated Notch signaling and enhanced IL-10 expression.
(A) BMDC were pre-treated with 10 μM DAPT or an equal volume of DMSO, for 30 min, and then stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS for 24 h. IL-10 and IL-2 mRNA transcript levels were measured by qRT-PCR while IL-12 protein concentration was measured by ELISA. (B) BMDC were pre-treated with 10 μg/ml cycloheximide or an equal volume of H2O for 30 min and then stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS for 4 h. Relative mRNA transcript levels were measured by qRT-PCR. (C)Wild-type (WT) and IL-10−/− BMDC were stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS for 24 h and cytokine concentration was measured by ELISA. Data from all three parts are mean ± SD of triplicates and are representative of at least 3 independent experiments. Two-way ANOVA with Bonferroni post-test was used to statistically compare stimuli; P<0.05 was considered significant. * = P<0.05, ** = P<0.01, *** = P<0.001, ns = not significant. ND = not detected.
To determine whether IL-10 and IL-2 are direct targets of this non-canonical Notch signaling pathway, BMDC were stimulated with Jagged1-Fc and/or LPS in the presence of cycloheximide and IL-10 and IL-2 mRNA transcript levels were quantified after 4 h (Figure 3B). Only Jagged1-Fc induced IL-10 was insensitive to cycloheximide – indicating that Notch signaling in response to Jagged1-Fc can directly induce expression of IL-10 transcripts. LPS induced IL-2 was prevented by cycloheximide, indicating that TLR induced IL-2 expression is dependent on de novo protein synthesis. Similarly, Jagged1-Fc induced IL-2, as well as IL-10 and IL-2 transcripts induced by combined Jagged1-Fc and LPS stimulation were completely abolished by cycloheximide, suggesting that interaction between the Notch and TLR signaling pathways requires de novo protein synthesis for enhanced IL-10 and IL-2 expression.
As IL-10 can act via an autocrine mechanism to inhibit IL-12 expression (25, 26), BMDC from IL-10 deficient mice were stimulated with Jagged1-Fc and LPS. As expected IL-10 deficiency had no effect on secretion of IL-2 in response to either LPS or Jagged1-Fc or both, while LPS induced IL-12 was significantly enhanced by IL-10 deficiency (Figure 3C). However, IL-10 deficiency did not prevent suppression of LPS induced IL-12 by Jagged1-Fc – indicating that enhanced IL-10 expression is not sufficient to explain inhibition of LPS induced IL-12 expression mediated by non-canonical Notch signaling.
In summary Notch and TLR signaling interact in DC to modulate DC responses to TLR ligands via a non-canonical Notch signaling pathway and this interaction is dependent on de novo protein synthesis.
PI3K signaling is required for modulation of IL-10 and IL-2 secretion by Notch and TLR cross-talk in DC
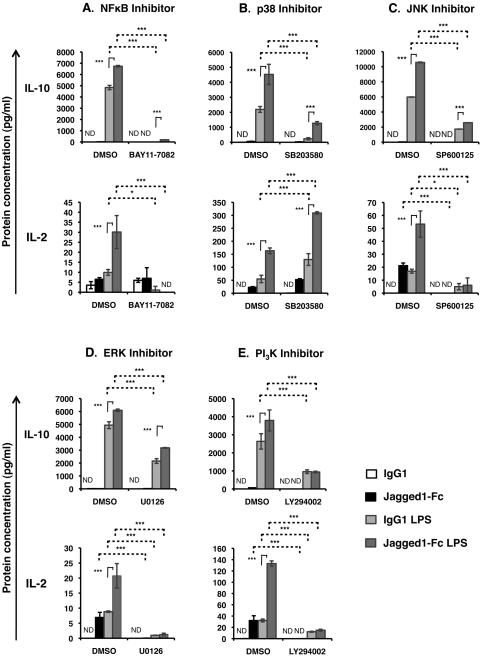
As Notch and TLR signaling could interact through either the MyD88 or TRIF dependent pathways we focused on downstream signaling molecules that are common to both pathways, in particular NFκB, the MAPKs and PI3K. To establish which of these downstream signaling molecules are required for interactions between Notch and TLR signals in DC, BMDC were stimulated with Jagged1-Fc and/or LPS in the presence or absence of inhibitors of each of these. Inhibition of NFκB, p38, JNK, ERK or PI3K signaling resulted in reduced IL-10 in response to LPS, and in response to combined Jagged1-Fc and LPS stimulation (Figure 4). However, compared to LPS acting alone, combined Notch and TLR4 signaling still enhanced IL-10 expression in the presence of the NFκB, p38, JNK, or ERK inhibitors. Only inhibition of PI3K signaling with LY294002 prevented Notch mediated enhancement of LPS induced IL-10 expression (Figure 4E), indicating that PI3K signaling is required for the interaction between the Notch and TLR pathways that leads to enhanced IL-10 expression while signaling through NFκB and the MAPKs is dispensable. Levels of Jagged1-Fc induced IL-10 expression were too low to reliably determine the effect of these inhibitors on Jagged1-Fc induced IL-10 expression.
Figure 4. PI3K activity is required for Jagged1-Fc mediated enhancement of both LPS induced IL-10 and IL-2.
BMDC were pre-treated with (A) BAY11-7082, (B) SB203580, (C) SP600125, (D) U0126, (E) LY294002 or a corresponding quantity of vehicle control (DMSO) for 1 hour, and then stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS. Accumulation of IL-10 and IL-2 in the supernatant after 4-24 h was measured by ELISA (A - 8 h, B and C - 24, D - 8, and E - 4). Data in all parts are mean ± SD of triplicates and are representative of at least 3 independent experiments. Two-way ANOVA with Bonferroni post-test was used to statistically compare stimuli; P<0.05 was considered significant. * = P<0.05, ** = P<0.01, *** = P<0.001, ns = not significant. ND = not detected.
Inhibition of PI3K, ERK and JNK significantly inhibited expression of IL-2 in response to either LPS or Jagged1-Fc or both acting together (Figure 4C-E), demonstrating that PI3K, ERK and JNK are all necessary for enhanced IL-2 expression in response to combined Notch and TLR signaling. Inhibition of NFκB signaling had no effect on Jagged1-Fc induced IL-2 expression but completely inhibited expression of IL-2 in response to LPS and Jagged1-Fc + LPS (Figure 4A), indicating that NFκB activity is crucial for IL-2 in response to TLR signaling. In contrast, inhibition of p38 activity lead to enhanced IL-2 expression in response in response to either Notch or TLR4 stimulation, indicating that p38 signaling acts to limit IL-2 expression (Figure 4B). However, the ability of combined Notch and TLR stimulation to enhance IL-2 expression by 2 fold or more was not affected by the p38 inhibitor suggesting that p38 is not involved in cross-talk between the pathways.
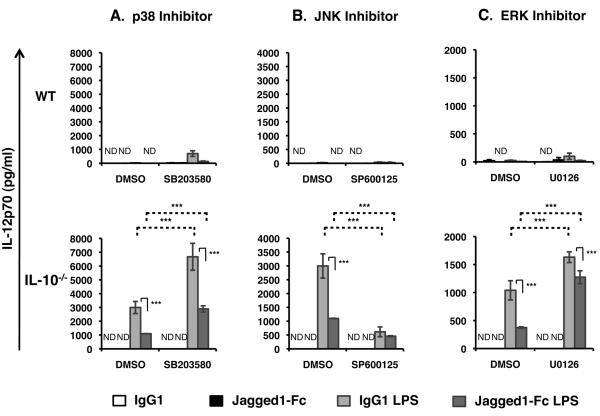
As wild-type BMDC make only relatively low levels of IL-12 in response to LPS it was not possible to reliably determine whether inhibition of NFκB, p38, JNK, ERK or PI3K signaling affected IL-12 secretion in response to combined Jagged1-Fc + LPS stimulation. Instead BMDC derived from IL-10−/− mice were used as these DC produce a much higher level of IL-12 following LPS stimulation. Inhibition of p38 or ERK signaling enhanced both LPS induced and Jagged1-Fc + LPS induced IL-12 (Figure 5A and C). However, inhibition of neither p38 nor ERK altered Notch mediated suppression of LPS induced IL-12, suggesting these signaling molecules are dispensable for modulating IL-12 expression. Unfortunately, it was not possible to determine the effect of PI3K inhibition on IL-12 expression as IL-12 starts accumulating in the supernatant after 8 hours by which time a significant level of cell toxicity was observed in cultures containing LY294002. However, stimulating BMDC in the presence of the JNK inhibitor SP600125 mimicked the effect of Notch on LPS induced IL-12 – inhibiting IL-12 secretion by around 6 fold (Figure 5B). Combined Jagged1-Fc + LPS stimulation was unable to further inhibit IL-12 in the presence of the JNK inhibitor, suggesting that Notch may act to inhibit LPS induced IL-12 by inhibiting JNK signaling.
Figure 5. LPS induced IL-12 expression requires JNK activity.
BMDC from wild-type (WT) or IL-10−/− mice were pre-treated with (A) SB203580, (B) SP600125, (C) U0126, or a corresponding quantity of vehicle control (DMSO) for 1 hour, and then stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS. Concentration of IL-12 in the supernatant after 8 (C) or 24 h (A and B) was measured by ELISA. Data are mean ± SD of triplicates and are representative of at least 3 independent experiments. Two-way ANOVA with Bonferroni post-test was used to statistically compare stimuli; P<0.05 was considered significant. *** = P<0.001, ns = not significant. ND = not detected.
In contrast to modulation of cytokine secretion, modulation of Hes1 expression by combined Notch and TLR stimulation was not affected by inhibition of NFκB, p38, JNK, ERK or PI3K signaling (Supplementary Figure S1C-G).
PI3K is activated by Notch signaling in a γ-secretase independent manner
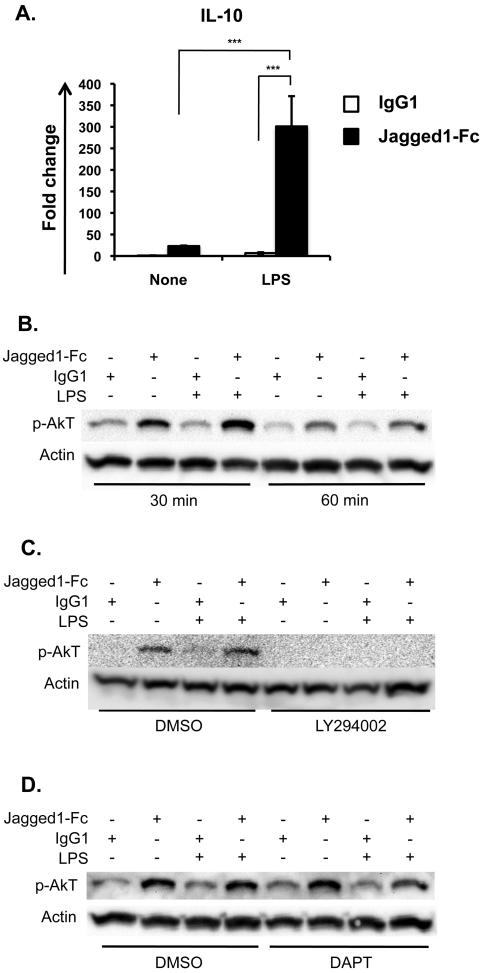
Activation of PI3K leads to phosphorylation of Akt, one of its downstream targets. To establish whether Notch signaling can activate PI3K directly we analysed phosphorylation of Akt in the DC like cell line JAWSII by western blot. Jagged1-Fc and LPS both induced IL-10 mRNA transcripts in JAWSII and IL-10 transcript levels were greatly enhanced when both stimuli were provided simultaneously (Figure 6A). Western blot confirmed increased phosphorylation of Akt in cells treated with the Notch ligand Jagged1-Fc (Figure 6B), and that phosphorylation of Akt in response to Jagged1-Fc was dependent on PI3K activity (Figure 6C). Interestingly, phosphorylation of Akt was independent of γ-secretase activity (Figure 6D). All together this demonstrates that non-canonical Notch signalling can activate PI3K directly.
Figure 6. Jagged1-Fc can activate Akt in the presence of γ-secretase inhibitors.
(A) JAWSII were stimulated with Jagged1-Fc (black bars) or IgG1 (white bars) in the presence or absence of 100 ng/ml LPS for 3 h and IL-10 transcripts were measured by qRT-PCR. (B) JAWSII were stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS and phosphorylated Akt was detected by western blot after 30 or 60 min. (C) JAWSII were pre-treated with LY294002 or vehicle control for 30 min and then stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS and phosphorylated Akt was detected by western blot after 30 min. (D) JAWSII were pre-treated with DAPT or vehicle control for 30 min and then stimulated with Jagged1-Fc or IgG1 in the presence or absence of 100 ng/ml LPS and phosphorylated Akt was detected by western blot after 30 min. Data in (A) are mean ± SD of triplicates and all data are representative of 3 independent experiments. Two-way ANOVA with Bonferroni post-test was used to statistically compare stimuli; P<0.05 was considered significant. * = P<0.05, ** = P<0.01, *** = P<0.001, ns = not significant. ND = not detected.
Discussion
DC derived cytokines play a key role in specifying T helper cell differentiation and thus the type of immune response mounted following detection pathogenic material. The cytokine signature of a DC depends on not only on the activating pathogen derived molecules sensed by the DC but also on the local environment present during activation. It is not yet known how environmental cues affect DC maturation but it is clear that immune responses show tissue specificity as pathogens in the skin elicit different immune responses compared with those present in the blood or affecting the gut. As Notch signaling is known to influence cell fate decisions and Notch ligands are expressed in a tissue specific manner all over the body we were interested in whether Notch signaling can act as an endogenous environmental signal and influence DC maturation to pathogen derived stimuli. We demonstrate here that the Notch and TLR pathways interact in DC, and as a result of this interaction DC stimulated simultaneously with Notch and TLR ligands have a distinct cytokine profile, characterized by enhanced IL-10 and IL-2 and reduced IL-12 expression, compared to DC stimulated with either Notch or TLR ligands alone. This interaction between Notch and TLR signalling occurs through a non-canonical γ-secretase independent Notch signalling pathway and is dependent on PI3K signalling.
PI3K can be activated downstream of TLR signaling via direct interaction with either MyD88 or TRIF (27, 28). PI3K activity leads to downstream signaling through activation of Akt and Bruton’s tyrosine kinase (Btk). Both Akt and Btk enhance NFκB activity via phosphorylation of the p65 subunit (29, 30). However, inhibitor and knock-out (p85α) studies have demonstrated that PI3K signaling also acts to limit expression of pro-inflammatory modulators in response to TLR activation. In particular PI3K signaling is associated with altering TLR responses to enhance IL-10 and reduce IL-12 expression, via both IL-10 dependent and independent mechanisms (31, 32). Modulation of IL-10 and IL-12 expression by PI3K signaling is likely to involve Akt and modulation of MAPK activity via interactions with the MAPKKKs (MAP Kinase Kinase Kinase) (32, 33). As concurrent Notch and TLR signals lead to enhanced IL-10 and reduced IL-12 through IL-10 dependent and independent mechanisms, we speculated that PI3K could be involved in the interaction between these two pathways. Using phosphorylation of Akt as a read-out for PI3K activity, we show that γ-secretase independent Notch signaling in DC activates PI3K. Inhibition of PI3K with LY294002 further confirmed that increased Akt phosphorylation in response to Jagged1-Fc corresponds to increased PI3K activity.
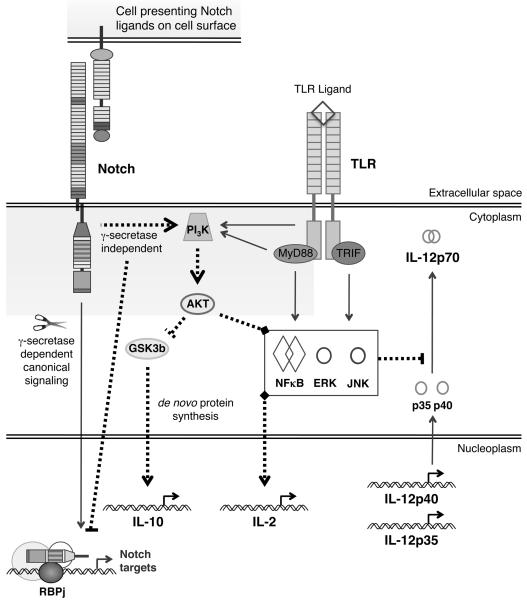
Consistent with our findings a number of studies have shown that Notch signaling activates the PI3K-Akt pathway in a variety of cell lines (12, 34-38). Furthermore, Sade et al. (38) showed Notch and PI3K could physically interact in Jurkats over-expressing N1IC and Perumalsamy et al. (12) demonstrated that Notch1 promoted Akt activation in Hela cells via a non-canonical route, independent of RPBj. It was not clear whether γ-secretase cleavage was required for activation of Akt, however, the authors did show that Akt activation required a cytoplasmic localization for Notch and that a membrane tethered NIC fragment was sufficient to induce phosphorylation of Akt, suggesting that activation of the PI3K-Akt pathway may occur in the absence of γ-secretase mediated cleavage. Thus, combined with our findings that Notch acts to modulate TLR induced cytokine expression and that this critically requires PI3K activity, we propose that Notch alters the DC cytokine profile by activating PI3K and thus Akt, which modulates the activity of NFκB and the MAPKs (Figure 7).
Figure 7. Proposed mechanism for Notch and TLR interaction in DC.
Our data support a model whereby non-canonical Notch signaling and both MyD88 dependent and TRIF dependent TLR signalling interact through convergence on PI3K and Akt, which then alters the cytokine profile of these cells by modulating the activities of GSK3β, NKκB, ERK and JNK to enhance transcription of the IL-10 and IL-2 genes, as well as inhibiting generation of biologically active IL-12p70.
Modulation of IL-10, IL-2 and IL-12 expression occurs through three distinct routes. Enhanced IL-10 and IL-2 expression occurs at the transcriptional level and relies on de novo synthesis of at least one unknown protein, while inhibition of IL-12 occurs post-transcriptionally – most likely by preventing the post-translational modifications required for IL-12 secretion. Inhibition of JNK activity mimicked Notch mediated repression of IL-12 secretion, suggesting Notch signaling via PI3K-Akt may interfere with post-transcriptional regulation of IL-12 by regulating JNK activity. While NFκB, p38, JNK, ERK and PI3K activity are all required for optimal IL-10 expression in response to a TLR stimulus, only PI3K activity was required for enhanced IL-10 expression in response to simultaneous Notch and TLR ligation. Enhanced IL-2 expression, however, required ERK, JNK and NFκB activity in addition to PI3K activity. As Akt signaling can modulate the activity of ERK, JNK, and NFκB, it is likely that non-canoncial Notch signaling enhances TLR induced IL-2 expression through activation of PI3K and Akt (Figure 7).
Interaction between the Notch and TLR pathways was bidirectional – in addition to modulation of TLR induced cytokines, mRNA transcript levels of the canonical Notch target gene Hes1 were suppressed. Suppression of Hes1 mRNA following simultaneous Notch and TLR stimulation did not require NFκB, ERK, JNK, p38 or PI3K activity, suggesting that inhibition of Hes1 may reflect competition between the canonical and alternative Notch signaling pathways rather than direct inhibition by TLR signaling. Studies reporting putative pathways for non-canonical Notch signaling support a model where canonical and alternative Notch signaling pathways compete with one another as the protein-protein interactions and/or localizations required are often mutually exclusive. For example Song et al. (39) showed that Akt can phosphorylate NIC, which promotes localization to the cytoplasm and results in reduced expression of canonical Notch targets as less NIC is available to bind to RBPj in the nucleus. These data combined with the finding that Notch requires a cytoplasmic localization in order to active Akt (12) suggests that as Notch enhances Akt activation, Akt in turn promotes a cytoplasmic localization for Notch that further enhances this alternative signaling pathway through activation of Akt to the detriment of canonical Notch target expression in the nucleus. Thus the reduction in Hes1 expression reported here may simply indicate that in the presence of a TLR stimulus non-canonical Notch pathway predominates over canonical signaling. Competition between canonical and alternative Notch pathways may help explain apparently contradictory data published by Hu et al. (22) suggesting that TLR signaling promotes expression of Hes1 in human monocytes incubated with M-CSF for 24 h in a γ-secretase and RBPj dependent manner, as whether the canonical or the alternative pathway predominates will depend not only on TLR signaling but also on the cell type and differentiation stage. Indeed, two microarray studies following differentiation of human monocytes to macrophages showed that Hes1 is relatively highly expressed in monocytes but is down regulated during differentiation and is expressed only at low levels in mature macrophages (40, 41). This suggests that canonical Notch signaling is strong in monocytes relative to that in fully differentiated macrophages or DC. We hypothesize that under conditions of strong canonical Notch signaling, such as in monocytes, combined TLR and Notch signaling is insufficient to favour the alternative Notch pathway and allow it to outcompete the dominant canonical pathway. In mature cells, by contrast, where canonical signaling appears to be reduced, the canonical and alternative pathways are more evenly balanced and the addition of TLR signaling allows the alternative pathway to predominate. In support of our data with DC, other studies have suggested Hes1 expression is inhibited by TLR signaling in mature primary alveolar macrophages and rat microglial cells as well as in the macrophage cell line RAW264.7 (42-44).
Competition between pathways could suggest the involvement of different Notch receptors. However, while APCs predominantly express the Notch2 receptor and a lower level of Notch1 mRNA transcripts (Supplementary Figure S2), activation with LPS has only a relatively modest effect on overall Notch receptor transcript levels (14, 42, 44-47 and data not shown).
Notch ligation has been shown to induce DC maturation, although these Notch conditioned DC exhibit a distinct cytokine profile and altered T cell stimulatory capacities when compared to DC induced by TLR ligands (16, 17, 48). However, little is known how Notch signaling in DC may affect responses to TLR ligands. Other studies have suggested that Notch and TLR pathways may interact in macrophages, however, contradictory data have been published regarding the outcome of any such interactions. In two studies Notch signaling in response to recombinant Jagged1-Fc or over-expressed N1IC in microglial cells or RAW267.4 resulted in reduced inducible nitric oxide synthase (iNOS) and pro-inflammatory cytokine expression in response to LPS (42, 44). Other studies using a γ-secretase inhibitor and over-expressed N1IC suggested that canonical Notch signaling promoted TNFα, IL-6 and iNOS expression in RAW264.7 cells by promoting activation of NFκB (49, 46). However, these data must be interpreted carefully as over-expression of N1IC results in an intracellular concentration of N1IC many orders of magnitude greater than required for signaling and thus may allow interactions that could not be possible under physiological conditions, and γ-secretase inhibitors prevent cleavage of more than 25 proteins in addition to Notch (50). We demonstrate there are two routes for Notch signaling in DC, and that these two pathways compete. Thus while our data does not exclude the possibility that canonical Notch signaling may interact with TLR induced NFκB activity as suggested above it may explain these otherwise contradictory data as, if the alternative and canonical pathways each have opposing effects on TLR signaling, differences in culture conditions and method for inducing or inhibiting Notch signaling will affect which pathway predominates and thus the outcome of combined Notch and TLR signaling. Differential roles for canonical versus non-canonical Notch signaling in DC could be further investigated using the recently generated conditional Nicastrin knockout (51). Nicastrin is a critical component of the γ-secretase complex and thus the conditional knockout would allow study of the non-canonical Notch signaling pathway in isolation.
Notch signaling is known to influence cell fate decisions in response to external stimuli and this work demonstrates that a concurrent Notch signal can modulate the functional maturation of DC to stimulation with a TLR ligand, in terms of the secretion of immunostimulatory cytokines. This altered cytokine profile will affect the outcome of DC-T cell interactions and the resulting adaptive immune response. Jagged1 is expressed on the surface of many different cell types but it is not currently known what cells are capable of delivering a Notch signal to DC, and thus it is not known where and to what purpose Notch signaling may regulate DC maturation to TLR ligation in vivo. Due to the fact that Jagged1 knock-out is embryonic lethal and it is not possible to reliably administer immobilized Notch ligands to mice in a fashion which can be shown to induce Notch signaling in the relevant cell type, our in vitro data cannot be supported by in vivo evidence at this time. Further investigation will be required to confirm the biological relevance of these findings in the whole organism. However, we hypothesize that expression of Jagged ligands may act as an endogenous environmental cue that acts to fine-tune the functional maturation of DC to complement the environment in which TLR ligation occurred. Differential expression of Jagged ligands may help explain why different tissues can induce subtly different adaptive responses to the same microbial stimulus. The intestine is an example of a tissue or organ where immune responses must be tightly regulated in order to prevent development of excessive inflammation. Both regulatory T cells and expression of IL-10 and IL-2 have been shown to be important for limiting inflammation in the gut, while excessive TH1 and TH17 responses cause chronic intestinal inflammation. Thus through enhancing expression of IL-10 and IL-2 whilst limiting IL-12, Notch signaling may play an important role in modulating immune responses to TLR stimuli in order to maintain gut integrity and homeostasis. While Notch and TLR cross-talk in DC may be beneficial in some tissues, this interaction may lead to or facilitate disease when Jagged ligands are over-expressed out of context. For example, a relatively high level of Jagged1 expression is associated with aggressive breast and prostate cancers (52, 53) and Jagged1 may act to facilitate tumour progression through altering the functional maturation of DC and subsequent immune responses.
Understanding how Notch and TLR signaling pathways interact in order to modulate the functional maturation of DC to inflammatory stimuli may guide the development of new approaches for manipulating DC in the treatment of diseases resulting from a disproportional immune response such as in autoimmunity, cancer and infectious disease.
Supplementary Material
Acknowledgements
We thank Emily Kay and Brendan Wren (London School of Hygiene and Tropical Medicine) for providing the C. jejuni cultures, Gerry Weinmaster (UCLA) for providing the L cell lines and Ana Rosario (NIMR, London) for the providing IL-10−/− mice with permission from the MGC Foundation, Germany and Neil Rogers and Caetano Reis e Sousa (Cancer Research UK) for providing MyD88−/− and TRIF−/− mice with permission from Shizuo Akira, Osaka University.
Footnotes
This work was supported by the Wellcome Trust, Grant number 078366/Z/05/A.
Abbreviations used in this paper: BMDC - Bone marrow-derived dendritic cell; DAPT - N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; DC - Dendritic cell; C. jejuni - Campylobacter jejuni; NIC - Intracellular domain of Notch; qRT-PCR - Quantitative Real-Time PCR; RBPj: recombination signal binding protein for immunoglobulin kappa J; TRIF - TIR-domain-containing adapter-inducing interferon-β.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Joffre O, Nolte MA, Sporri R, Reis e Sousa C. Inflammatory signals in dendritic cell activation and the induction of adaptive immunity. Immunol Rev. 2009;227:234–247. doi: 10.1111/j.1600-065X.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 5.Dallman MJ, Smith E, Benson RA, Lamb JR. Notch: control of lymphocyte differentiation in the periphery. Curr Opin Immunol. 2005;17:259–266. doi: 10.1016/j.coi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berechid BE, Kitzmann M, Foltz DR, Roach AH, Seiffert D, Thompson LA, Olson RE, Bernstein A, Donoviel DB, Nye JS. Identification and characterization of presenilin-independent Notch signaling. J Biol Chem. 2002;277:8154–8165. doi: 10.1074/jbc.M108238200. [DOI] [PubMed] [Google Scholar]
- 10.Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, Grigsby PW, Miner JH, Farr AG, Kopan R. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez Arias A, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr Opin Genet Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- 12.Perumalsamy LR, Nagala M, Banerjee P, Sarin A. A hierarchical cascade activated by non-canonical Notch signaling and the mTOR-Rictor complex regulates neglect-induced death in mammalian cells. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.20. [DOI] [PubMed] [Google Scholar]
- 13.Ramain P, Khechumian K, Seugnet L, Arbogast N, Ackermann C, Heitzler P. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr Biol. 2001;11:1729–1738. doi: 10.1016/s0960-9822(01)00562-0. [DOI] [PubMed] [Google Scholar]
- 14.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi E, Chiba S, Kumano K, Kunisato A, Takahashi T, Hirai H. Expression of Notch ligands, Jagged1, 2 and Delta1 in antigen presenting cells in mice. Immunol Lett. 2002;81:59–64. doi: 10.1016/s0165-2478(01)00326-1. [DOI] [PubMed] [Google Scholar]
- 16.Bugeon L, Gardner LM, Rose A, Gentle M, Dallman MJ. Cutting Edge: Notch Signaling Induces a Distinct Cytokine Profile in Dendritic Cells That Supports T Cell-Mediated Regulation and IL-2-Dependent IL-17 Production. J Immunol. 2008;181:8189–8193. doi: 10.4049/jimmunol.181.12.8189. [DOI] [PubMed] [Google Scholar]
- 17.Weijzen S, Velders MP, Elmishad AG, Bacon PE, Panella JR, Nickoloff BJ, Miele L, Kast WM. The Notch ligand Jagged-1 is able to induce maturation of monocyte-derived human dendritic cells. J Immunol. 2002;169:4273–4278. doi: 10.4049/jimmunol.169.8.4273. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 19.Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 20.Carra G, Gerosa F, Trinchieri G. Biosynthesis and posttranslational regulation of human IL-12. J Immunol. 2000;164:4752–4761. doi: 10.4049/jimmunol.164.9.4752. [DOI] [PubMed] [Google Scholar]
- 21.Lyakh L, Trinchieri G, Provezza L, Carra G, Gerosa F. Regulation of interleukin-12/interleukin-23 production and the T-helper 17 response in humans. Immunol Rev. 2008;226:112–131. doi: 10.1111/j.1600-065X.2008.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, Tateya T, Kang YJ, Han J, Gessler M, Kageyama R, Ivashkiv LB. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friis LM, Keelan M, Taylor DE. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect Immun. 2009;77:1553–1560. doi: 10.1128/IAI.00707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rathinam VA, Appledorn DM, Hoag KA, Amalfitano A, Mansfield LS. Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through TLR4-MyD88 and TLR4-TRIF axes. Infect Immun. 2009 doi: 10.1128/IAI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aste-Amezaga M, Ma X, Sartori A, Trinchieri G. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J Immunol. 1998;160:5936–5944. [PubMed] [Google Scholar]
- 26.Huang LY, Reis e Sousa C, Itoh Y, Inman J, Scott DE. IL-12 induction by a TH1-inducing adjuvant in vivo: dendritic cell subsets and regulation by IL-10. J Immunol. 2001;167:1423–1430. doi: 10.4049/jimmunol.167.3.1423. [DOI] [PubMed] [Google Scholar]
- 27.Aksoy E, Vanden Berghe W, Detienne S, Amraoui Z, Fitzgerald KA, Haegeman G, Goldman M, Willems F. Inhibition of phosphoinositide 3-kinase enhances TRIF-dependent NF-kappa B activation and IFN-beta synthesis downstream of Toll-like receptor 3 and 4. Eur J Immunol. 2005;35:2200–2209. doi: 10.1002/eji.200425801. [DOI] [PubMed] [Google Scholar]
- 28.Ojaniemi M, Glumoff V, Harju K, Liljeroos M, Vuori K, Hallman M. Phosphatidylinositol 3-kinase is involved in Toll-like receptor 4-mediated cytokine expression in mouse macrophages. Eur J Immunol. 2003;33:597–605. doi: 10.1002/eji.200323376. [DOI] [PubMed] [Google Scholar]
- 29.Arbibe L, Mira JP, Teusch N, Kline L, Guha M, Mackman N, Godowski PJ, Ulevitch RJ, Knaus UG. Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat Immunol. 2000;1:533–540. doi: 10.1038/82797. [DOI] [PubMed] [Google Scholar]
- 30.O’Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity. 2008;29:12–20. doi: 10.1016/j.immuni.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Fukao T, Tanabe M, Terauchi Y, Ota T, Matsuda S, Asano T, Kadowaki T, Takeuchi T, Koyasu S. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol. 2002;3:875–881. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- 32.Ohtani M, Nagai S, Kondo S, Mizuno S, Nakamura K, Tanabe M, Takeuchi T, Matsuda S, Koyasu S. Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood. 2008;112:635–643. doi: 10.1182/blood-2008-02-137430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukao T, Koyasu S. PI3K and negative regulation of TLR signaling. Trends Immunol. 2003;24:358–363. doi: 10.1016/s1471-4906(03)00139-x. [DOI] [PubMed] [Google Scholar]
- 34.Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 35.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 36.Mungamuri SK, Yang X, Thor AD, Somasundaram K. Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006;66:4715–4724. doi: 10.1158/0008-5472.CAN-05-3830. [DOI] [PubMed] [Google Scholar]
- 37.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, Bhagat G, Agarwal AM, Basso G, Castillo M, Nagase S, Cordon-Cardo C, Parsons R, Zuniga-Pflucker JC, Dominguez M, Ferrando AA. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem. 2004;279:2937–2944. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- 39.Song J, Park S, Kim M, Shin I. Down-regulation of Notch-dependent transcription by Akt in vitro. FEBS Lett. 2008;582:1693–1699. doi: 10.1016/j.febslet.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Shi B, Huang CC, Eksarko P, Pope RM. Transcriptional diversity during monocyte to macrophage differentiation. Immunol Lett. 2008;117:70–80. doi: 10.1016/j.imlet.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 42.Grandbarbe L, Michelucci A, Heurtaux T, Hemmer K, Morga E, Heuschling P. Notch signaling modulates the activation of microglial cells. Glia. 2007;55:1519–1530. doi: 10.1002/glia.20553. [DOI] [PubMed] [Google Scholar]
- 43.Kim MY, Park JH, Mo JS, Ann EJ, Han SO, Baek SH, Kim KJ, Im SY, Park JW, Choi EJ, Park HS. Downregulation by lipopolysaccharide of Notch signaling, via nitric oxide. J Cell Sci. 2008;121:1466–1476. doi: 10.1242/jcs.019018. [DOI] [PubMed] [Google Scholar]
- 44.Monsalve E, Perez MA, Rubio A, Ruiz-Hidalgo MJ, Baladron V, Garcia-Ramirez JJ, Gomez JC, Laborda J, Diaz-Guerra MJ. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J Immunol. 2006;176:5362–5373. doi: 10.4049/jimmunol.176.9.5362. [DOI] [PubMed] [Google Scholar]
- 45.Fung E, Tang SM, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, Carlesso N, Aster JC, Aikawa M. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 46.Palaga T, Buranaruk C, Rengpipat S, Fauq AH, Golde TE, Kaufmann SH, Osborne BA. Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur J Immunol. 2008;38:174–183. doi: 10.1002/eji.200636999. [DOI] [PubMed] [Google Scholar]
- 47.Sekine C, Moriyama Y, Koyanagi A, Koyama N, Ogata H, Okumura K, Yagita H. Differential regulation of splenic CD8- dendritic cells and marginal zone B cells by Notch ligands. Int Immunol. 2009;21:295–301. doi: 10.1093/intimm/dxn148. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Cabezas B, Naranjo-Gomez M, Bastos-Amador P, Requena-Fernandez G, Pujol-Borrell R, Borras FE. Ligation of notch receptors in human conventional and plasmacytoid dendritic cells differentially regulates cytokine and chemokine secretion and modulates th cell polarization. J Immunol. 2011;186:7006–7015. doi: 10.4049/jimmunol.1100203. [DOI] [PubMed] [Google Scholar]
- 49.Monsalve E, Ruiz-Garcia A, Baladron V, Ruiz-Hidalgo MJ, Sanchez-Solana B, Rivero S, Garcia-Ramirez JJ, Rubio A, Laborda J, Diaz-Guerra MJ. Notch1 upregulates LPS-induced macrophage activation by increasing NF-kappaB activity. Eur J Immunol. 2009;39:2556–2570. doi: 10.1002/eji.200838722. [DOI] [PubMed] [Google Scholar]
- 50.Parks AL, Curtis D. Presenilin diversifies its portfolio. Trends Genet. 2007;23:140–150. doi: 10.1016/j.tig.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 51.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, Liu C, Ibrahim S, Beran M, Zavadil J, Efstratiadis A, Taghon T, Michor F, Levine RL, Aifantis I. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 53.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, Rubin MA, Aster JC. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.