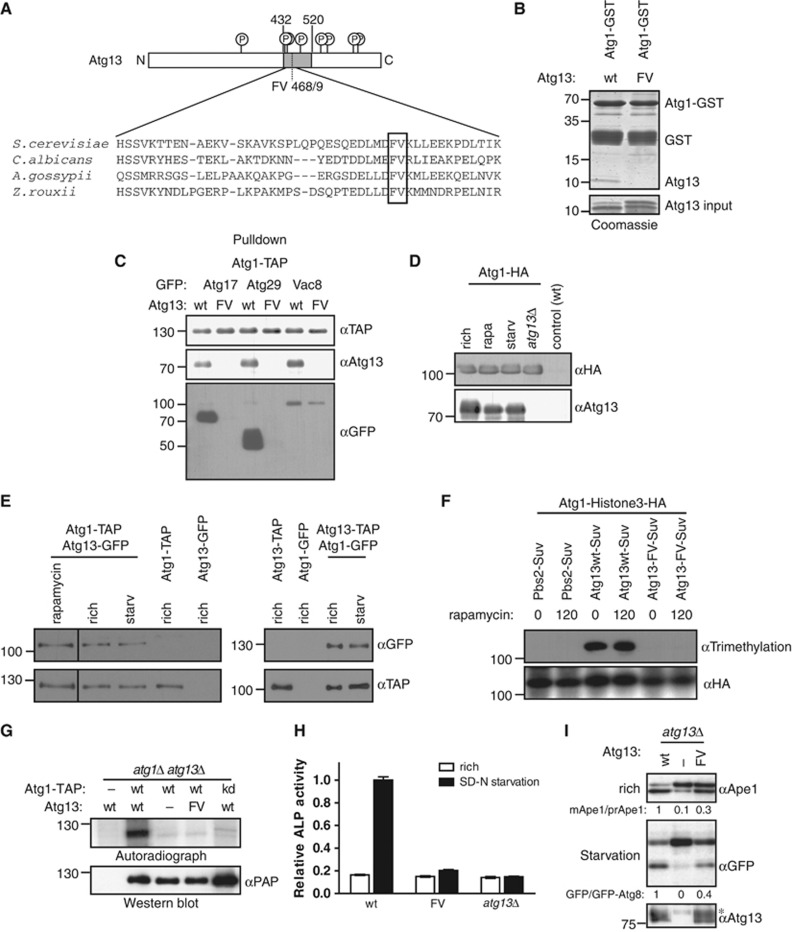
Figure 1.
Atg13 binding to Atg1 promotes Cvt and autophagy function, but is not regulated by starvation conditions. (A) Schematic representation of budding yeast Atg13 and alignment of the Atg1-binding region with homologues from different yeast species. The residues required for Atg1 binding are marked with a grey box. Circled ‘P’ mark known phosphorylation sites on Atg13. N: amino-terminus; C: carboxy-terminus. (B) A fragment encompassing amino acids 432–520 of Atg13 (wt) or the corresponding FV mutant (FV) was expressed in E. coli and incubated in excess with an immobilized GST-tagged fragment of Atg1 (amino acids 501–897) purified from E. coli (GST-Atg1). Input (5.5%) and bound proteins were analysed by coomassie blue staining, which stains proteins proportional to their size. The size of marker proteins (in kDa) is indicated on the left. (C) ATG1-TAP atg13Δ cells containing endogenously GFP-tagged Atg17, Atg29 or Vac8 and wild-type (wt) or the Atg13 FV-mutant (F468A; V469A) were grown to mid log phase. Atg1 was immunoprecipitated and its association with Atg13 and the GFP-tagged proteins was analysed by immunoblotting. Extract inputs are shown in Supplementary Figure S1B. (D) atg1Δ and atg1Δatg13Δ cells expressing HA-tagged Atg1 and wild-type (wt) cells containing an empty plasmid were grown to mid log phase (rich), and then treated with rapamycin (rapa) or starved for 4 h in SD-N medium (starv). Atg1 was immunoprecipitated and its association with Atg13 analysed by immunoblotting. Note that phosphorylated Atg13 isolated from cells grown in rich medium migrates slightly slower on SDS–PAGE, and the extent corresponds to the shift typically observed under our gel conditions. (E) Yeast cells containing endogenously tagged Atg1-TAP with and without endogenously tagged Atg13-GFP, or conversely cells containing endogenously tagged Atg13-TAP and Atg1-GFP expressed from a centromeric plasmid were grown to mid log phase (rich) and treated with rapamycin (rapa) or starved for 4 h in SD-N medium (starv). Atg1-TAP and Atg13-TAP were affinity purified and their association with GFP-tagged proteins was analysed by western blotting. (F) Histone3-HA tagged Atg1 was expressed together with wild-type (wt) or the Atg13-FV mutant fused to Suv methylase (Suv) in atg1Δatg13Δ cells, or for control together with Pbs2-Suv in atg1Δ cells. Logarithmically growing cultures were treated with rapamycin (rapa) for 0 or 120 min, and methylation was monitored by preparing cell extracts and western blotting with an anti-trimethylation-specific antibody. Note that Suv-, GFP- or TAP-tagged Atg13 are fully functional, however, do not show the characteristic phosphorylation-induced reduced mobility observed with the untagged protein under our gel conditions. (G) atg1Δatg13Δ cells containing TAP-tagged wild-type Atg1 (wt) or the Atg1 kinase-dead (kd) K54A mutant and wild-type Atg13, the Atg13-FV mutant or an empty plasmid were grown to mid log phase. Atg1 was immunoprecipitated and its autophosphorylation activity was measured by autoradiography in vitro. (H) pho8Δ60pho13Δatg13Δ cells expressing wild-type Atg13 (wt), the Atg13-FV mutant (FV) or an empty plasmid were grown to mid log phase and starved for 4 h in SD-N medium. Pho8Δ60-specific alkaline phosphatase (ALP) activity (nmol/min/mg) was measured in three independent experiments as described in ‘Materials and methods’, and plotted as relative ALP activity with standard deviation (s.d.) compared to the wild-type controls. (I) atg13Δ cells expressing GFP-Atg8 and containing wild-type Atg13, the Atg13-FV mutant or an empty plasmid were grown to mid log phase with and without starvation for 4 h in SD-N. Processing of endogenous Ape1 and cleavage of GFP-Atg8 was analysed by western blotting, and quantified by calculating the ratio of cleaved versus uncleaved Ape1 or GFP-Atg8, respectively. The asterisk marks a non-specific band detected by the Atg13 antibody. Figure source data can be found with the Supplementary data.