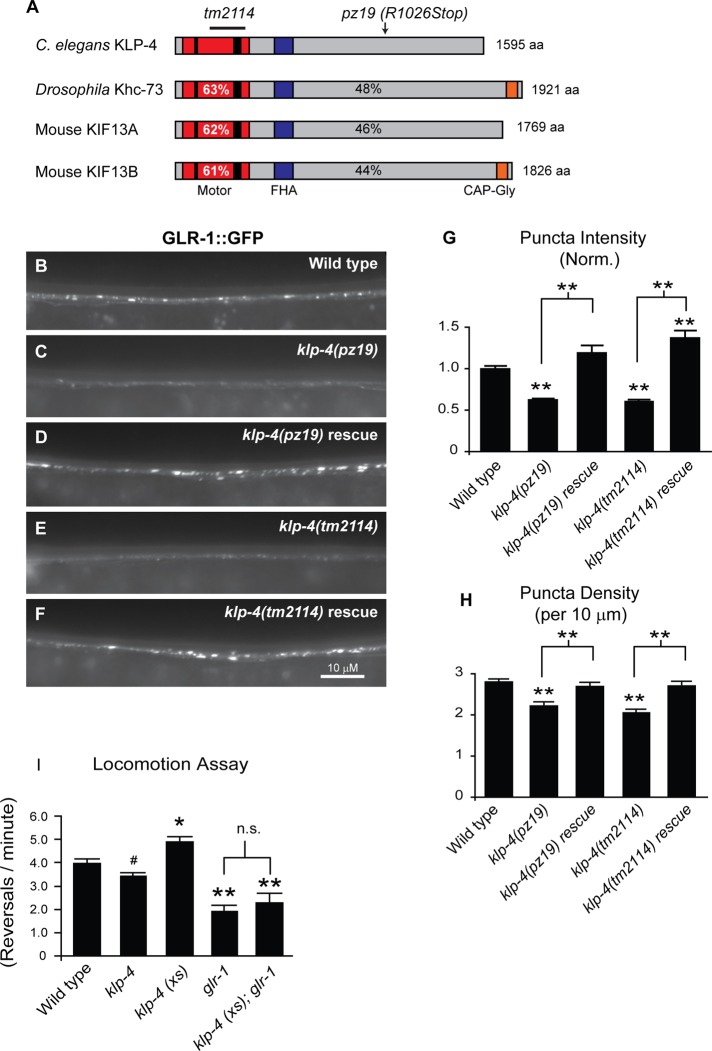
FIGURE 1:
The kinesin KLP-4 functions in the VNC to regulate the abundance of GLR-1 and GLR-1–dependent behavior. (A) Protein domain organization of C. elegans KLP-4, Drosophila Khc-73, and mouse KIF13A and KIF13B. The N-terminal motor domain (red box) containing the signature ATP-binding motif (thin black bar) and microtubule-binding regions (thick black bar), the cargo-binding domain (gray box), the Forkhead-associated domain (FHA; purple box), and the microtubule plus end–binding domain (CAP-Gly; orange box) are shown. The percentage identity of the motor domain of C. elegans KLP-4 (percentages in white) or full-length KLP-4 (percentages in black) vs. Drosophila Khc73 and mouse KIF13A and KIF13B are indicated. (B–F) Representative images of the anterior VNC of larval stage 4 (L4) wild-type (B), klp-4(pz19) (C), rescued klp-4(pz19) (D), klp-4(tm2114) (E), and rescued klp-4(tm2114) (F) animals expressing an integrated GLR-1::GFP transgene (nuIs25). In these and all subsequent images anterior is to the left and ventral is up. (G, H) Quantification of GLR-1::GFP puncta intensities (normalized; G) and densities (H) for the strains pictured in B–F. Means and SEM are shown for n = 38 wild-type, n = 23 klp-4(pz19), n = 21 rescued klp-4(pz19), n = 20 klp-4(tm2114), and n = 21 rescued klp-4(tm2114) animals. (I) Quantification of number of spontaneous reversals per minute for n = 20 wild-type, n = 20 klp-4(tm2114), and n = 23 animals expressing an integrated klp-4 transgene under the control of the glr-1 promoter (klp-4(xs)) (pzIs20), n = 14 glr-1(n2461), and n = 13 glr-1(n2461) animals overexpressing klp-4 (klp-4(xs)). Values that differ significantly from wild type are indicated by asterisks above each bar, whereas other comparisons are marked by brackets (**p ≤ 0.001, *p ≤ 0.01, #p ≤ 0.05, Tukey–Kramer test).