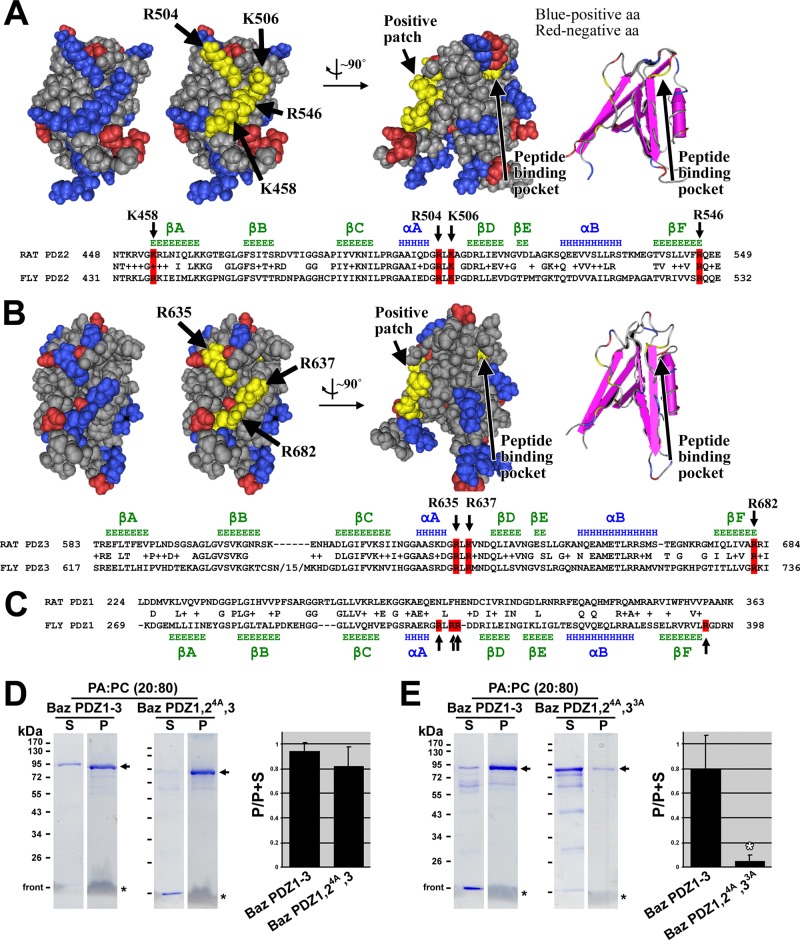
FIGURE 4:
Mapping residues on Bazooka PDZ domains responsible for phosphatidic acid binding. (A) A Cn3D space-filling representation of the solution structure of rat Par-3 PDZ2 (Molecular Modeling Database [MMDB] ID: 61497; Protein Data Bank [PDB] ID: 2OGP) shows negatively charged residues in red and positively charged residues in blue. A positively charged patch is highlighted in yellow, and the residues involved are indicated with arrows. Rotation of the structure by ∼90° shows the relative position of the peptide-binding pocket (the residue at the base of the pocket is highlighted in yellow, and the pocket is also shown with secondary structures, far right). A sequence alignment between rat Par-3 PDZ2 and Baz PDZ2 is shown below, with conserved residues of the positive patch highlighted in red. Secondary structure data above the alignment are based on the structure of rat Par-3 PDZ2 (β-strands in green and α-helices in blue; Wu et al., 2007). (B) A Cn3D space-filling representation of the solution structure of rat Par-3 PDZ3 (MMDB ID: 64814; PDB ID: 2K1Z) with negatively charged residues in red and positively charged residues in blue. A positively charged patch is highlighted in yellow, and the residues involved are indicated with arrows. Rotation of the structure by ∼90° shows the relative position of the peptide-binding pocket (the residue at the base of the pocket is highlighted in yellow, and the pocket is also shown with secondary structures, far right). A sequence alignment between rat Par-3 PDZ3 and Baz PDZ3 is shown below, with conserved residues of the positive patch highlighted in red. Secondary structure data above the alignment are based on the structure of rat Par-3 PDZ3 (β-strands in green and α-helices in blue; Feng et al., 2008). (C) A sequence alignment between rat Par-3 PDZ1 and Baz PDZ1. Secondary structure data below the alignment are based on secondary structure predictions of Baz PDZ1 using Jpred3, HNN, Prof, SOOPMA, and PORTER secondary structure prediction tools (β-strands in green and α-helices in blue). Positively charged residues found at similar positions as those in Baz PDZ2 and 3 are indicated in red. These are not conserved in rat Par-3 PDZ1. (D) Mutation of the four positive amino acid residues in PDZ2 to alanine in Baz PDZ1-3 has no effect on PA liposome binding. (E) Mutation of the four positive amino acid residues in PDZ2 plus the three positive amino acid residues in PDZ3 to alanine in Baz PDZ1-3 abrogates PA liposome binding. The experiments in D and E were conducted in the same way as those in Figure 2.