Background: The Mre11 complex plays an important role in DNA DSB repair.
Results: The Rad50 zinc hook domain is critical for the recruitment of the Mre11 complex to chromosomal DSBs.
Conclusion: The Rad50 zinc hook domain is important for initiating DNA damage responses and mediating DSB repair.
Significance: These studies help to understand how the Mre11 complex functions to repair DNA lesions and to maintain genome stability in mammalian cells.
Keywords: DNA Damage Response, DNA Repair, Genomic Instability, Homologous Recombination, Mammal, ATM Activation, DNA Damage Check Points, DNA Double-stranded Breaks (DSBs), Rad50 Zinc Hook, Mre11-Rad50-Nbs1 (MRN) Complex
Abstract
The Mre11-Rad50-Nbs1 (MRN) complex plays critical roles in checkpoint activation and double-stranded break (DSB) repair. The Rad50 zinc hook domain mediates zinc-dependent intercomplex associations of MRN, which is important for DNA tethering. Studies in yeast suggest that the Rad50 zinc hook domain is essential for MRN functions, but its role in mammalian cells is not clear. We demonstrated that the human Rad50 hook mutants are severely defective in various DNA damage responses including ATM (Ataxia telangiectasia mutated) activation, homologous recombination, sensitivity to IR, and activation of the ATR pathway. By using live cell imaging, we observed that the Rad50 hook mutants fail to be recruited to chromosomal DSBs, suggesting a novel mechanism underlying the severe defects observed for the Rad50 hook mutants. In vitro analysis showed that Zn2+ promotes wild type but not the hook mutant of MR to bind double-stranded DNA. In vivo, the Rad50 hook mutants are defective in being recruited to chromosomal DSBs in both H2AX-proficient and -deficient cells, suggesting that the Rad50 hook mutants are impaired in direct binding to chromosomal DSB ends. We propose that the Rad50 zinc hook domain is important for the initial binding of MRN to DSBs, leading to ATM activation to phosphorylate H2AX, which recruits more MRN to the DSB-flanking chromosomal regions. Our studies reveal a critical role for the Rad50 zinc hook domain in establishing and maintaining MRN recruitment to chromosomal DSBs and suggest an important mechanism of how the Rad50 zinc hook domain contributes to DNA repair and checkpoint activation.
Introduction
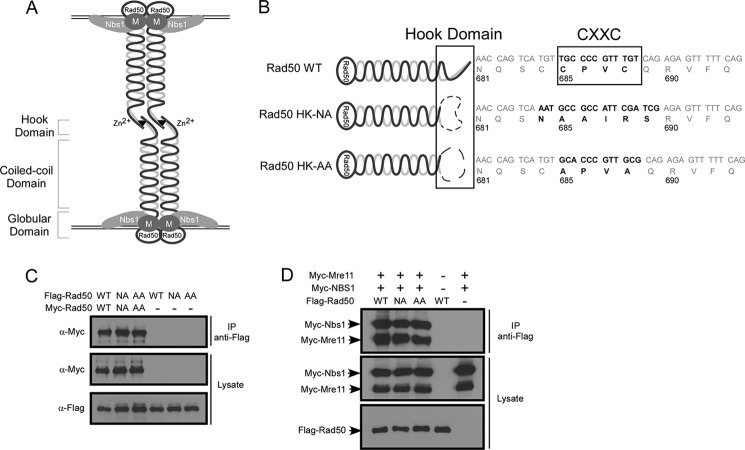
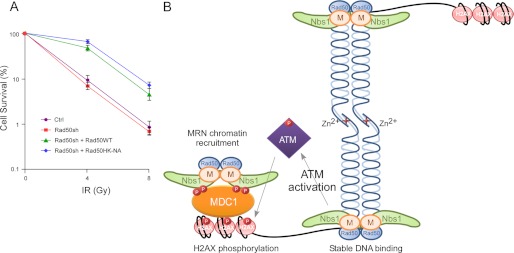
MRN2 plays important roles in various DNA damage responses, contributing to the maintenance of genome stability (1–3). MRN binds to DNA DSBs and is required for ATM activation to initiate the checkpoint signaling cascade. It is also critical for DNA end processing to initiate homologous recombination (HR)-mediated DSB repair (4). Mre11 (M) and Rad50 (R) are the core of the complex, and structural analysis revealed that MR forms a heterotetramer (M2R2) (5). The Mre11 dimer and the two Rad50 ATPase domains form a globular DNA-binding domain, and Nbs1 joins the complex by directly interacting with Mre11 (6). The coiled-coil domain of Rad50 protrudes from the globular domain, and at its apex, a conserved CXXC motif described as a hook interacts with a second zinc hook motif from another Rad50 molecule through a coordinating Zn2+ ion (7) (depicted in Fig. 1A).
FIGURE 1.
The Rad50 zinc hook domain is important for tethering of the MRN complex. A, schematic representation of the architecture of the MRN complex and its interaction with DNA. Rad50 forms a homodimer and interacts with Mre11 and NBS1 through the globular domain, forming a DNA-binding motif. At the apex of the coiled-coil domain, the Rad50 zinc hook domain links with another Rad50 zinc hook domain, resulting in the tethering of DNA ends or sister chromatids. B, a schematic drawing of Rad50 hook mutants generated. For the Rad50HK-NA mutant, the CXXC and adjacent two amino acids were replaced with the NAAIRS flexible polylinker. For the Rad50HK-AA mutant, the two conserved cysteine residues at amino acids 685 and 688 were replaced with alanines. C, dimer formation of Rad50 hook mutants. Myc-tagged or FLAG-tagged Rad50 WT and the hook mutants Rad50HK-AA or Rad50HK-NA were co-expressed in 293T cells. Co-immunoprecipitation was carried out from whole cell lysates with anti-FLAG agarose beads, and Western blotting was performed with indicated antibodies. D, interaction of Rad50 hook mutants with Mre11 and Nbs1. Myc-tagged Mre11 and Nbs1 were co-expressed with FLAG-tagged Rad50WT or hook mutants Rad50HK-AA and Rad50HK-NA in 293T cells. Whole cell lysate was subjected to Co-IP by anti-FLAG antibody, followed by Western blotting using the indicated antibodies. IP, immunoprecipitation.
Studies in yeast demonstrate that the Rad50 zinc hook domain is important for various Mre11 complex functions including DSB repair, telomere maintenance, and meiotic DSB formation (8). The phenotype of the Rad50 hook mutants is as severe as Rad50-null mutants, suggesting that the Rad50 zinc hook domain is essential for MRN functions in yeast. In vitro analysis showed that MRN binds and tethers DNA ends (9, 10). Atomic force microscopy further revealed that DNA binding induces conformational change of the coiled-coil domain of Rad50, favoring intercomplex interactions of M2R2 through the zinc hook domain of Rad50, which promotes DNA tethering (11). In yeast, live cell imaging revealed that the two ends of a DSB are held together to prevent chromosome breakage and ensure correct repair of DNA breaks (12, 13). In this aspect, the yeast Mre11 complex and especially the Rad50 zinc hook contribute to the holding of chromosomal DSB ends, suggesting that the DNA tethering function of Rad50 is important for preventing chromosome breakage. However, in mammalian cells, it was suggested that MRN does not play a significant role in the holding of broken chromosome ends (14), which is different from the findings in yeast.
Although severe defects of Rad50 hook mutants were found in yeast (8), its role in mammalian cells was not clear. In this study, we showed that the Rad50 hook mutants fail to activate ATM, exhibit defects in end resection and ATR activation, and are impaired in HR-mediated DSB repair in mammalian cells. We further demonstrated that the Rad50 zinc hook domain is essential for the recruitment of MRN to chromosomal DSBs, providing a mechanistic explanation for the severe defects observed in the Rad50 hook mutants for checkpoint activation and DSB repair. In addition, we showed that zinc-dependent intercomplex associations of MRN through the zinc hook domain of Rad50 stabilize MRN binding to DNA, thereby promoting stable interaction of MRN with chromosomal DSBs to activate ATM. Failure in ATM-mediated phosphorylation of H2AX in Rad50 hook mutants further impairs MRN chromatin binding at DSB-flanking regions through the γH2AX-dependent pathway. Our studies thus reveal critical biological functions of the Rad50 zinc hook domain in mammalian cells and identify a new role of the Rad50 zinc hook domain in localizing MRN to chromosomal DSBs to initiate DNA damage responses.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, Retroviral Infection, and Stable Cell Line Generation
U2OS and 293T cells were cultured in DMEM supplemented with 10% FBS and antibiotics at 37 °C with 5% CO2. Insect cell line Sf21 was cultured in Grace's insect medium supplemented with 10% FBS and antibiotics at 25 °C. Insect cell line Sf9 was cultured in SF900 II SFM medium (Invitrogen) at 27 °C. U2OS and 293T cells were transfected by CaCl2. To generate FLAG-tagged, Myc-tagged, and EGFP-tagged Rad50, Mre11, and Nbs1 and HA-Mdc1 stable cell lines, U2OS cells were first transfected with pcDNA3-based or EGFP-C1/N1-based plasmids expressing exogenously tagged proteins, followed by G418 or puromycin selection.
Plasmids and shRNA
To generate the Rad50 hook mutants Rad50HK-NA and Rad50HK-AA (see Fig. 1B), site-directed mutagenesis was conducted by QuikChange mutagenesis (Stratagene). Oligonucleotides 5′-ACAGACGAAAACCAGTCAAATGCCGCCATTCGATCGAGAGTTTTTCAGACAGAG-3′ and 5′-GAAAACCAGTCATGTGCACCCGTTGCGCAGAGAGTTTTTCAG-3′ were used as primers for Rad50HK-NA and Rad50HK-AA mutants, respectively. The shRNA-resistant wild type Rad50 and Rad50 hook mutants were constructed by mutating four nucleotides at the shRNA targeting sequences by site-directed mutagenesis using the primer 5′-ACCAGAATTAAGAAATAAGCTTCAGAATGTCAATAGA-3′.
Gene silencing was performed by using pMKO-based retroviral infection (15). The shRNA oligonucleotides were annealed and inserted into the pMKO retroviral vector. The shRNA sequences used were for Rad50, GAAACAAACTGCAGAATGTCACTC, and for H2AX, GGGACGAAGCACTTGGTAACA. pMKO-EBFP-puro was constructed by inserting the coding region of enhanced blue fluorescence protein (EBFP) (Addgene plasmid 14893) (16) in front of puromycin open reading frame to generate a fusion EBFP-puro marker in pMKO vectors containing shRNA target sequences.
Expression and Purification of Recombinant Proteins, DNA Binding Assay
Recombinant baculoviruses expressing FLAG- or Myc-tagged Rad50 (wild type or hook mutants), Nbs1, and Mre11 were generated by using the Bac-to-Bac baculovirus expression system (Invitrogen). To express MRN complex or the Mre11 and Rad50 complex (MR), His-tagged FLAG-Rad50 (wild type or the hook mutant NA), Mre11 and/or Nbs1 were co-expressed in Sf9 cells by baculovirus infection, and the complex was purified as described (17).
For the DNA binding assay, a 5′ forward oligonucleotide primer was biotinylated and used with unlabeled reverse primer DNA fragments (0.5 kb) for PCR amplification of the plasmid pUC19. 100 or 250 ng of these 0.5-kb DNA fragments were incubated with 10 μl of M-270 streptavidin Dynabeads (Invitrogen) for each reaction in the buffer containing PBS with 0.1% Tween 20. Purified MR or MRN wild type or hook mutant complexes were exchanged to EDTA-containing buffer (50 mm HEPES, pH 7.5, 50 mm KCl and 5 mm MgCl2, 100 mm EDTA) or Zn2+-containing buffer (50 mm HEPES, pH 7.5, 50 mm KCl and 5 mm MgCl2, 150 mm zinc acetate). After washing DNA-binding beads with either with EDTA- or Zn2+-containing buffer, MR or MR(HK-NA) complexes were incubated with DNA containing beads in either EDTA- or Zn2+-containing buffer for 1.5 h at 4 °C. After washing, the samples containing DNA beads were boiled with 2× SDS loading buffer and detected by immunoblotting for Mre11 and Rad50.
Antibodies, Immunofluorescence, and Western Blotting
The antibodies used include anti-Myc (9E10; Abcam), anti-FLAG-M2 (Sigma), anti-HA-11 (Covance), anti-Ku70 (Santa Cruz Biotechnology), anti-Chk1-S317p, anti-Chk2-T68p, anti-H2AX (Bethyl), anti-H2AX-S139p, and anti ATM-S1981p (Cell Signaling Technology), anti-Rad50 (Novus Biologicals), anti-RPA2 (CalBiochem), anti-Nbs1 (18), and anti-Mre11 (19).
For immunofluorescence analysis, the cells were cultured on sterile coverslips. After whole cell gamma-IR or laser-induced microirradiation, the cells were fixed at indicated time points with 4% paraformaldehyde for 10 min, permeabilized in 0.5% Triton X-100 for 10 min, blocked with 3% BSA in PBS for 30 min, and incubated with indicated primary antibodies and FITC-conjugated secondary antibody (Jackson Immunoresearch). Cellular nuclei were marked by Hoechst staining. A Nikon Eclipse E800 fluorescence microscope and SPOT Advanced imaging software were used for analysis of fluorescence microscopy images.
Immunoprecipitation and Western blot analysis were performed as described (20, 21). The cells were lysed with NETN buffer (20 mm Tris, pH 8.0, 1 mm EDTA, 150 mm NaCl, and 0.5% Nonidet P-40) for 30 min. Cleared cell lysates were then collected for immunoprecipitation or boiled in 2× SDS loading buffer and subjected to SDS-PAGE.
Laser Microirradiation and Live Cell Imaging
DNA DSBs were generated in live cells expressing EGFP-tagged Rad50 (wild type and hook mutants) and mRFP-Nbs1 by laser microirradiation using a picosecond short-pulsed green laser (a diode-pumped second harmonic 532-nm Nd:YAG laser microbeam with a 76-MHz repetition rate, 12-ps pulse duration) coupled to a Zeiss Axiovert fluorescence microscope (Laboratory of Dr. Michael W. Berns, University of California at San Diego) (22). Time lapse, live cell imaging was performed to monitor real time recruitment of fluorescent-tagged proteins to laser-induced damage sites. To quantitate recruitment, the absolute intensities were determined by using Image J software (National Institutes of Health) to measure the fluorescence signal intensities at the laser-induced damage sites, and the cellular background fluorescence intensities were subtracted. The data shown represent the averages of over 10 independent cell measurements, with error bars representing standard deviations.
Chromatin Isolation
Chromatin isolation was performed as described before (23). Briefly, the cells were washed with cold PBS and lysed in CSK buffer (10 mm PIPES, pH 6.8, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, 50 mm NaF, 0.1 mm sodium orthovanadate, 0.1% Triton X-100, and protease inhibitors) on ice for 10 min. Cytoplasmic proteins supernatant were removed from nuclei after low speed centrifugation (1,300 × g for 5 min). The nuclei pellets were washed in CSK buffer then lysed in solution (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, and protease inhibitors). After centrifugation at 1,700 × g for 5 min, the pellets were resuspended in CSK buffer. 2× SDS loading buffer was added, and samples were boiled for 10 min.
Homologous Recombination Assay
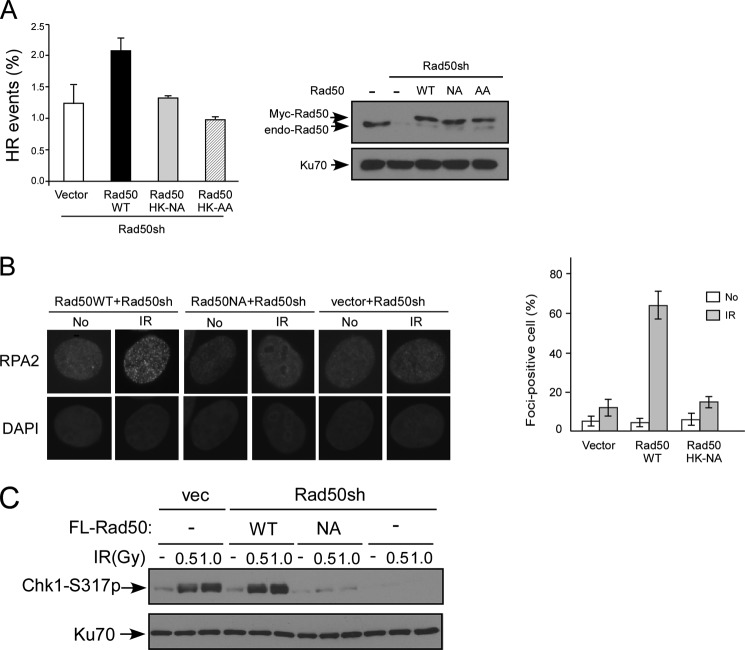
The EGFP-based HR repair substrate construct (supplemental Fig. S4) was described before (24). Briefly, the EGFP ORF with a 20-bp deletion in the middle was inserted with an I-Sce1 cleavage site, which is followed by an internal EGFP fragment with a truncated CMV promoter and truncated EGFP ORF (1–214 amino acids) (iEGFP). The EGFP HR repair substrate was stably integrated into U2OS cells, followed by hygromycin selection, and single clones were selected and confirmed by Southern blotting for single-copy integration (data not shown). U2OS EGFP HR cell line was first knocked down with Rad50 shRNAs and overexpressed with either Rad50 wild type or hook mutants. I-Sce1 was introduced by retroviral infection. FACS analysis was performed to assay for EGFP-positive, HR-repaired events.
RESULTS
The Rad50 Zinc Hook Domain Is Important for Recruiting MRN to DSBs on Chromatin
The conserved CXXC motif on Rad50 is critical for the zinc hook function to mediate the MR intercomplex interactions and DNA tethering (Fig. 1, A and B) (7). We generated the Rad50 hook mutants by mutating two conserved cysteine residues (amino acids 685 and 688) in the zinc hook motif to alanine (Rad50HK-AA) or by replacing six amino acids covering the zinc hook motif with a NAAIRS amino acid sequence (Rad50HK-NA), which is a flexible linker designed to minimize any potential conformational changes in the mutant protein (Fig. 1B) (25). We expressed Myc-tagged Rad50 WT and the hook mutants Rad50HK-AA and Rad50HK-NA along with FLAG-tagged Rad50WT or hook mutant alleles in 293T cells, and co-immunprecipitation revealed that the Rad50 hook mutants do not have defects in forming Rad50 homodimers (Fig. 1C). The expression of Myc-Rad50 and the Rad50 hook mutants is comparable with endogenous Rad50 (supplemental Fig. S1A). Furthermore, when expressed in 293T cells or U2OS cells, FLAG-tagged Rad50WT or the hook mutants Rad50HK-AA and Rad50HK-NA binds to Myc-tagged Mre11 and Nbs1 or endogenous Mre11 and Nbs1 at similar levels as shown by co-immunoprecipitation, suggesting that the Rad50 hook mutants retain normal function to form the MRN complex (Fig. 1D, supplemental Fig. S1B, and data not shown). We also performed similar co-immunoprecipitation experiments in the presence of ethidium bromide (EB, 400 μm), which selectively inhibits DNA and protein interactions (19, 26), and found that the Rad50 hook mutant Rad50HK-NA binds to Mre11 and Nbs1 to the same extent as wild type Rad50 (supplemental Fig. S1C). This suggests that the Rad50 hook mutant can efficiently assemble into MRN complex even without interacting with DNA. We further showed that mutating the Rad50 zinc hook domain does not change the interaction of MRN complex with BRCA1 (supplemental Fig. S1D), suggesting that the Rad50 hook mutant forms complex with Mre11 and Nbs1 properly without causing significant conformation change, and MRN containing the Rad50 hook mutant binds normally with BRCA1 (20).
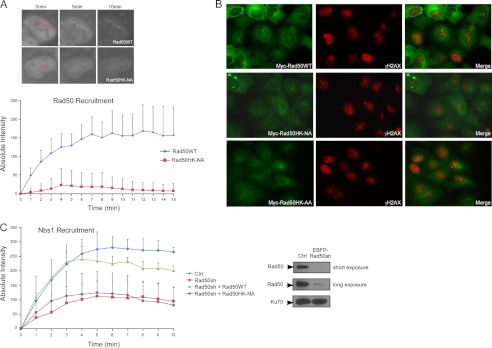
Upon DNA damage, MRN is recruited to DSBs to activate ATM and initiate DSB repair (27–29). To examine the recruitment of the Rad50 hook mutants to DSBs on chromatin, we used laser-induced microirradiation and monitored real time Rad50 recruitment in live cells. EGFP-tagged Rad50WT or the hook mutant Rad50HK-NA was transiently expressed in U2OS cells, and the recruitment of Rad50 proteins to DNA damage sites was followed under fluorescent microscopy. Although EGFP-Rad50WT is recruited to microlaser-treated sites quickly and efficiently, EGFP-Rad50HK-NA exhibits very limited recruitment (Fig. 2A and supplemental Fig. S2A). Similar results were obtained when we expressed Myc-Rad50WT or the hook mutants Myc-Rad50HK-NA and Myc-Rad50HK-AA in U2OS cells and performed immunostaining using an anti-Myc antibody after microirradiation (Fig. 2B). To examine whether Nbs1 and Mre11 recruitment to DNA damage sites is also impaired with the Rad50 hook mutants, we expressed Myc-Rad50WT or Myc-Rad50HK-NA mutant alleles carrying the silent mutations resistant to Rad50 shRNAs in U2OS cells, followed by inactivation of endogenous Rad50 by shRNAs. Live cell microscopy analysis showed that mRFP-tagged Nbs1 is recruited to damage sites efficiently in control U2OS cells and Rad50WT-reconstituted cells (with endogenous Rad50 silenced), whereas the recruitment is significantly impaired in the Rad50HK-NA hook mutant to a similar extent as that of cells expressing Rad50 shRNAs without reconstitution (Fig. 2C). Similar results of EGFP-Mre11 recruitment were obtained in the Rad50 hook mutant cell lines (data not shown). These studies suggest that the Rad50 zinc hook domain is essential for the recruitment of the MRN complex to chromosomal DSBs.
FIGURE 2.
The Rad50 zinc hook domain is important for the recruitment of MRN to chromosomal DSBs. A, recruitment of Rad50 to DNA damage sites in live cells. U2OS cells expressing EGFP-Rad50WT or the hook mutant Rad50HK-NA were induced with DNA DSBs by laser microirradiation. Recruitment of EGFP-Rad50WT and EGFP-Rad50HK-NA to laser-microirradiated sites at indicated time points are shown (upper panel, with red lines showing the laser-induced damage path in precut cells), and the absolute intensities of Rad50 recruitment were determined (bottom panel). B, stable U2OS cell lines expressing Myc-tagged Rad50WT or Rad50 hook mutants Rad50HK-NA and Rad50HK-AA were subjected to laser microirradiation (in which damage path generated by the laser mircrobeam was made as two crossed lines in an X-shaped pattern) and fixed 10 min later, followed by immunostaining using anti-Myc and anti-γH2AX antibodies. C, Nbs1 recruitment in Rad50 hook mutant cells. mRFP-Nbs1 with either EGFP-Rad50WT or EGFP-Rad50HK-NA or vector were co-transfected into U2OS cells, with endogenous Rad50 silenced by shRNAs or mock treated (Ctrl). Rad50 shRNA was engineered into pMKO-EBFP-puro construct, which uses EBFP to indicate cells containing shRNAs. DNA damage was induced by laser microirradiation, and the recruitment intensities of mRFP-Nbs1 were determined. Western blot shows silencing of endogenous Rad50 by EBFP-marked Rad50 shRNAs, with Ku70 used as a loading control.
The Rad50 Zinc Hook Domain Is Important for H2AX-independent Binding of MRN to Chromosomal DSBs and Promoting ATM Activation
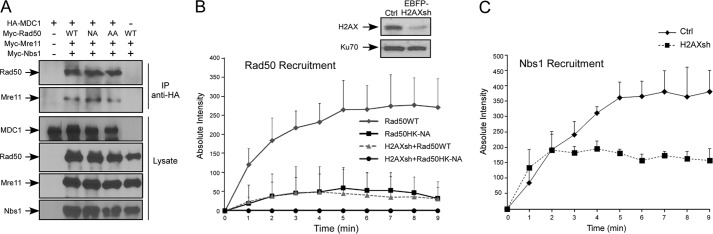
MRN is recruited to chromosomal DSBs by using two different mechanisms. One is by direct binding of MRN to DSB ends, which is γH2AX-independent, and the other is through localizing to DSB-surrounding chromatin regions through the interaction of Nbs1 with MDC1 in a γH2AX-dependent manner (30–35). To investigate the mechanisms underlying the severe defects of the Rad50 hook mutants localizing to chromosomal DSBs, we examined the interaction of MRN with MDC1 when the Rad50 zinc hook motif is mutated. We expressed Myc-Rad50WT or Myc-Rad50HK-NA along with Mre11 and Nbs1 and found that the interaction of MRN with FLAG-MDC1 is not affected by mutating the Rad50 zinc hook motif (Fig. 3A and data not shown). This study suggests that the chromosomal recruitment defects of the Rad50 hook mutants are not due to modulation of the interaction of MRN with MDC1, which mediates the recruitment of MRN to DSB-flanking chromatin.
FIGURE 3.
The Rad50 zinc hook domain is important for H2AX-independent binding of MRN to chromosomal DSBs. A, interaction of MRN complex with MDC1. Myc-tagged Mre11 and Nbs1 with either Myc-tagged Rad50WT or Rad50 hook mutants Rad50HK-AA and Rad50HK-NA was co-transfected with HA-MDC1 or control (−) in 293T cells. Anti-HA immunoprecipitation (IP) was carried out in whole cell lysates and followed by Western blotting with indicated antibodies. B, recruitment of Rad50 in H2AX knockdown cells. EGFP-Rad50WT or EGFP-Rad50HK-NA was transfected into U2OS cells, with endogenous H2AX silenced by EBFP-marked H2AX shRNAs. The absolute intensities of Rad50 recruitment to the damage sites after laser microirradiation were determined. Western blot shows silencing of endogenous H2AX, with Ku70 used as a loading control. C, recruitment of Nbs1 in H2AX-deficient cells. mRFP-Nbs1 was transfected in U2OS cells with either mock (Ctrl) or H2AX knockdown (H2AXsh) cells. DNA damage was introduced by laser microirradiation. Absolute intensities of Nbs1 recruitment were determined at the indicated time points.
To examine whether the Rad50 hook mutants have defects in direct binding with chromosomal DSBs, we monitored Rad50 recruitment to microlaser-generated DSBs in γH2AX-deficient cells. We generated a retroviral shRNA vector with EBFP-puro, which marks the shRNA-expressing cells with EBFP (supplemental Fig. S2B). As expected, the recruitments of wild type Rad50, Nbs1 and Mre11 were much reduced when H2AX is inactivated by shRNAs (Fig. 3, B and C, and data not shown), because of a lack of DSB-flanking chromatin recruitment (35). Furthermore, although EGFP-Rad50WT still retains low levels of recruitment through direct DSB binding in H2AX-deficient cells, the recruitment of EGFP-Rad50HK-NA mutant is completely abolished (Fig. 3B). This suggests that the zinc hook domain of Rad50 is important for initial γH2AX-independent recruitment of Rad50 to chromosomal DSBs through direct DNA binding.
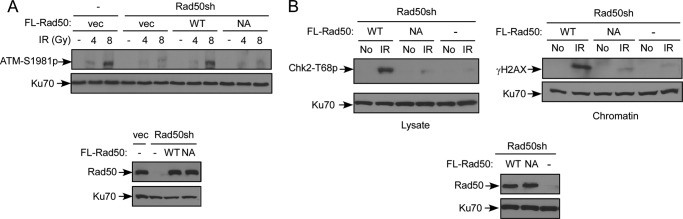
Because MRN is required for ATM activation, defective binding of the Rad50 hook mutant to DSBs prompted us to examine whether the Rad50 zinc hook domain is important for ATM activation. Myc-Rad50WT or the hook mutant Myc-Rad50HK-NA was expressed in U2OS cells with endogenous Rad50 inactivated by shRNAs. IR-induced ATM autophosphorylation and Chk2 phosphorylation at Thr-68 are both defective in the Rad50 hook mutants to a level similar as that of Rad50 knockdown cells without reconstitution (Fig. 4, A and B, left panel). We further examined H2AX phosphorylation at Ser-139 upon IR, and this ATM-dependent H2AX phosphorylation is also significantly reduced in the Rad50 hook mutant (Fig. 4B, right panel). These data suggest that the Rad50 hook mutant phenocopies the Rad50-deficient cells with severe defects in ATM activation and thus in phosphorylation of ATM substrates, which are likely due to impaired binding of the Rad50 hook mutant with chromosomal DSBs.
FIGURE 4.
The Rad50 zinc hook mutants are defective in ATM activation. A, the Rad50 zinc hook domain is important for ATM activation. U2OS cells expressing FLAG-tagged Rad50WT (WT), Rad50HK-NA (NA), or vector with mock (−) or endogenous Rad50 silenced by shRNAs. The cells were treated with IR (4 or 8 Gy) or no IR (−), and ATM autophosphorylation at Ser-1981 was examined by Western blot analysis (top panel). Expression levels of exogenously tagged Rad50WT or Rad50HK-NA mutant, with endogenous Rad50 silenced, are shown (bottom panel). Ku70 was used as a loading control. B, the Rad50 zinc hook domain is important for Chk2 phosphorylation and H2AX phosphorylation upon DNA damage. U2OS cells expressing FLAG-tagged Rad50WT (WT), Rad50HK-NA (NA), or vector were infected with Rad50 shRNAs. The cells were collected before (No) or after 1 h of IR treatment (8 Gy), and whole cell lysis (top left panel) and chromatin isolation (top right panel) were performed. Chk2 phosphorylation and γH2AX phosphorylation were examined by Western blot analysis using phospho-specific antibodies for Chk2-T68p and γH2AX-S139p. Expression levels of exogenously tagged Rad50WT or Rad50HK-NA mutant, with endogenous Rad50 silenced, are shown (bottom panel). Ku70 was used as a loading control.
Based on these observations, we proposed that the Rad50 hook mutants are impaired in the initial direct binding to DSBs, leading to compromised ATM activation. Because ATM-mediated H2AX phosphorylation is reduced, further localization of MRN containing the Rad50 hook mutant [MR(HK-NA)N] to DSB-flanking chromatin is also impaired, which results in severe recruitment defects of MR(HK-NA)N to chromosomal DSBs (see Fig. 7B).
FIGURE 7.
The Rad50 zinc hook domain is important for DSB repair because of its essential role in recruiting MRN to chromosomal DSBs. A, Rad50 hook mutant cells are sensitive to IR as revealed by clonogenic survival assay. U2OS cells expressing Rad50WT, Rad50HK-NA, or vector were mock-treated (Ctrl) or silenced for endogenous Rad50 by shRNAs. The cells were irradiated at the indicated doses, and the surviving colonies were counted after 2 weeks. The error bars represent the standard deviation from three independent experiments. B, model of MRN complex recruitment to the DSB ends. MRN complexes bind to the DSB ends and activate ATM, leading to γH2AX phosphorylation. Phosphorylated γH2AX activates MDC1, which recruits more MRN to the DSB-flanking chromatin. The Rad50 hook mutants are defective in ATM activation and downstream signaling cascade and thus fail to activate MRN complexes at DSB ends and to recruit more MRN complexes to DSB-flanking chromatin, resulting in DSB repair defects.
The Rad50 Hook Mutants Exhibit Defects in Zn2+-induced DNA Binding in Vitro
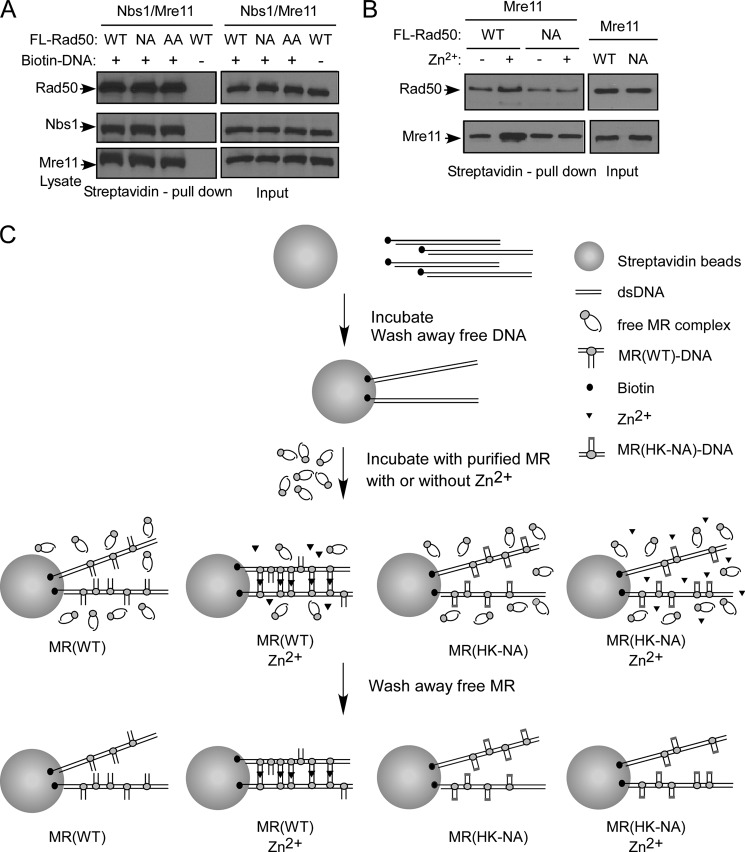
Because the Rad50 hook mutants show severe defects in direct association with chromosomal DSBs, we asked whether these Rad50 mutants are compromised for DNA binding when in complex with Mre11 and Nbs1. FLAG-tagged Rad50WT and the hook mutants Rad50HK-AA and Rad50HK-NA were expressed in insect cells by baculoviral infection along with Nbs1 and Mre11. We confirmed that the ability of Rad50 hook mutants to form Rad50 homodimers and the MRN complexes in insect cells is comparable with wild type Rad50 as revealed by co-immunoprecipitation (supplemental Fig. S3). We then performed DNA binding experiments using 0.5-kb double-stranded DNA (dsDNA) fragments with one end biotinylated and showed that the purified MRN complexes containing either wild type Rad50 or the hook mutants Rad50HK-AA or Rad50HK-NA bind to dsDNA at similar levels (Fig. 5A), suggesting that Rad50 hook mutants largely do not have defects in binding dsDNA with DSB ends.
FIGURE 5.
Rad50 zinc hook domain-mediated intercomplex interactions of MR promote more stable interaction of MRN with DNA in vitro in the presence of zinc. A, the MRN complexes containing wild type Rad50 or the Rad50 hook mutants bind to dsDNA at similar levels in the absence of zinc. MRN complexes containing FLAG-tagged Rad50WT or the Rad50 hook mutants FLAG-Rad50HK-NA and FLAG-Rad50HK-AA were expressed and purified from Sf9 insect cells after baculovirus infection. The MRN complexes (1.5 μg) were incubated with 250 ng of 0.5-kb biotinylated dsDNA fragments prebound to streptavidin Dynabeads. After washing, the binding of MRN with DNA was revealed by Western blot analysis using anti-FLAG, Mre11, and Nbs1 antibodies. B, Zn2+ promotes MR(WT) binding to streptavidin bead-bound DNA in vitro. 100 ng of biotinylated dsDNA fragments (0.5 kb) were incubated with streptavidin Dynabeads. After washing away free DNA, purified MR complexes (400 ng) containing FLAG-tagged Rad50WT or the Rad50 hook mutant Rad50HK-NA were incubated with DNA-bound streptavidin Dynabeads in the presence (+) or absence (−) of Zn2+. The binding of MR with DNA was revealed by Western blot analysis of Rad50 and Mre11 after washing away unbound free MR (also see C). C, the Rad50 hook-dependent intercomplex association may stabilize MRN to bind DNA. The addition of Zn2+ induces dsDNA-bound MR to form intercomplexes through the Rad50 zinc hook domains, resulting in more stable MR(WT) binding with dsDNA. This can be achieved by reduced disassociations of MR from DNA caused by MR intercomplex associations or increased affinity of MR for DNA binding induced by the conformational change upon MR intercomplex interactions through the Rad50 zinc hook domains. However, MR(HK-NA) complexes fail to form such Zn2+-dependent intercomplexes, and thus the dynamics of binding with dsDNA remains the same in the presence or absence of Zn2+.
In the in vitro analysis, the addition of Zn2+ induces the interaction between MRN complexes through the Rad50 zinc hook domain (7, 9, 11) (Fig. 1A). To examine whether the intercomplex association of MRN may stabilize MRN binding to DNA, MR complexes containing Rad50WT or the hook mutant Rad50HK-NA were purified from insect cells as described (17). Biotinylated 0.5-kb dsDNA fragments were bound to streptavidin Dynabeads. After washing away free DNA, streptavidin bead-bound DNA was incubated with wild type MR or MR(HK-NA). Interestingly, we found that the addition of Zn2+ increases the amounts of MR that is bound to a fixed number of dsDNA fragments attached to streptavidin beads (Fig. 5B). However, this Zn2+-dependent increase of DNA binding was not observed when the hook mutant Rad50HK-NA was used. This suggests that the MR intercomplex interaction through the zinc hook of Rad50 may promote more stable interactions of MR to DNA (Fig. 5C; see “Discussion” for more details), which contributes to the stable binding and the efficient recruitment of MRN to chromosomal DSBs.
The Rad50 Hook Mutants Are Defective in End Resection to Initiate HR and to Promote ATR-mediated Checkpoint Activation
To examine whether the Rad50 hook mutants have defects in HR-mediated DSB repair, we expressed Myc-tagged Rad50WT or the hook mutant Rad50HK-NA in U2OS cells carrying the EGFP-based HR substrate that is stably integrated in chromosomes (supplemental Fig. S4) (24). HR activity is examined by flow cytometry to determine the frequency of EGFP-positive, HR-mediated repair events after DSBs are generated by endonuclease I-Sce1. With the expression of endogenous Rad50 suppressed by shRNAs, HR in the Rad50 hook mutants Rad50HK-NA and Rad50HK-AA is impaired compared with that in Rad50 wild type cells (Fig. 6A). Furthermore, we found that end resection in the Rad50 hook mutants is much reduced by examining IR-induced RPA foci formation (Fig. 6B and supplemental Fig. S5), which reflects single-stranded DNA accumulation (36). Consistently, ATR activation as revealed by Chk1 phosphorylation is significantly compromised in the Rad50 hook mutants because of impaired accumulation of single-stranded DNA at DSB ends (Fig. 6C). In agreement with the defects of the Rad50 hook mutants in HR-mediated DSB repair and checkpoint activation, the loss of Rad50 zinc hook domain function causes IR-induced cell growth sensitivity (Fig. 7A).
FIGURE 6.
The Rad50 zinc hook mutants are defective in HR-mediated DSB repair and end resection. A, the Rad50 zinc hook domain is required for HR. U2OS cells with integrated EGFP HR repair substrate were stably expressed with Myc-tagged Rad50WT or hook mutants Rad50HK-AA and Rad50HK-NA, containing shRNA-resistant mutated sites, and endogenous Rad50 was silenced by shRNAs. The EGFP-positive cells representing the HR-repaired events were analyzed by FACS. Western blot shows expression of exogenously tagged Rad50WT and mutants and endogenous Rad50, with Ku70 used as a loading control. B, IR-induced RPA2 foci are reduced in the Rad50 hook mutant. U2OS cells expressing Rad50WT, Rad50HK-NA, or vector were silenced for endogenous Rad50 by shRNAs. RPA2 immunostaining was performed before (No) or 4 h after 10 Gy IR (IR), and RPA2 foci-positive cells were identified as cells with noticeable RPA2 foci formation (>30 foci/nuclei with bright, punctate staining) after IR (left), with DAPI staining of cellular nuclei. The percentage of cells containing RPA2 foci (>100 total cells counted for each experiment) of total number of cells counted is plotted (right), and the error bars represent the standard deviation from three independent experiments. C, IR-induced Chk1 phosphorylation is reduced in the Rad50 hook mutant. U2OS cells expressing FLAG-tagged Rad50WT, Rad50HK-NA, or vector (vec), with endogenous Rad50 silenced by shRNAs or mock (−), were treated with or without IR (5 or 10 Gy, 1 h after). Chk1 phosphorylation was monitored by phospho-specific antibody recognizing phosphorylated Ser-317. Ku70 was used as loading control.
DISCUSSION
The zinc hook domain of Rad50 is well conserved and mediates the intercomplex interactions of MRN through the apex of the coiled-coil arms of Rad50 (7, 8). In budding yeast, deleting the Rad50 zinc hook domain or mutating the critical amino acids in the zinc hook domain leads to severe defects in DNA DSB repair, telomere maintenance, and meiotic DSB formation (8). Based on these observations, it was suggested that the DNA tethering function or molecular bridging mediated by the Rad50 zinc hook domains is essential for DSB recombinational repair and other DNA damage responses (7–10). In this study, we observed that the Rad50 hook mutants are severely defective in localizing MRN to chromosomal DSBs in mammalian cells. This implicates a new role for the Rad50 zinc hook domain in the recruitment of MRN to chromosomal DSBs in addition to its DNA tethering function. Loss of DSB recruitment is likely the major cause for the severe defects observed in the Rad50 hook mutants in DSB repair, checkpoint activation, and other DNA damage responses.
Using live cell imaging, we monitored the recruitment of MRN to chromosomal DSBs and found that the Rad50 hook mutants fail to be localized to DSBs. It is well documented that MRN binds directly to DSBs, and this interaction is important for ATM activation (28, 29). It is also known that following initial ATM activation, ATM-dependent phosphorylation of H2AX triggers recruitment of more MRN to bind DSB-flanking chromatin, which is mediated by a direct binding of γH2AX with MDC1 and of MDC1 with the FHA and BRCT domains of Nbs1 (30–35). We showed that the interaction of MRN with MDC1 is not affected by mutating the Rad50 zinc hook domain, suggesting that the impaired localization of MRN to chromosomal DSBs is not due to loss of interactions of the MR(HK-NA)N mutants with MDC1. Instead, we found that ATM activation is abrogated in the Rad50 hook mutants as revealed by ATM autophosphorylation and Chk2 phosphorylation, and consequently H2AX phosphorylation is severely impaired. Loss of H2AX phosphorylation is expected to have dramatic effects on the recruitment of MRN to DSB-flanking chromatin (34, 35).
To understand how ATM activation is impaired when the zinc hook domain of Rad50 is mutated, we examined the recruitment of the Rad50 hook mutants to microlaser-generated DSB regions in H2AX-deficient cells. In addition to the recruitment of MRN to DSB-flanking chromatin regions, the Rad50 hook mutant is further defective in γH2AX-independent recruitment, suggesting that the hook mutant of MRN is indeed impaired in direct binding to chromosomal DSBs, which is likely a major cause for its failure in ATM activation. We thus propose that the hook mutant of MRN is impaired in the initial binding to chromosomal DSBs, which leads to defects in ATM activation. Because of impaired H2AX phosphorylation by ATM, MDC1 recruitment and the MDC1-dependent recruitment of MRN to DSB-flanking chromatin are compromised, thereby causing severe impairment of MRN localization to DSB-containing nuclear compartments (Fig. 7B).
To understand how the Rad50 hook mutants are impaired in chromosomal DSB binding, we performed in vitro analysis. We showed that the Rad50 hook mutant interacts with Nbs1 and Mre11 at similar levels as wild type Rad50. This is consistent with the observation in yeast that the interaction of Rad50 and Mre11 is not affected when the zinc hook domain of Rad50 is cleaved off by a protease in vivo (37). In addition, removing the zinc hook domain in yeast Rad50 does not have an effect on DNA binding as revealed by gel shift experiments (37). Similarly, we found that DNA binding activity of the Mre11 complex is largely unaffected when the Rad50 zinc hook domain is mutated. However, the addition of Zn2+ promotes more MR to bind DNA, but this effect was not observed when the Rad50 hook mutant was used. This suggests that zinc-dependent MR intercomplex interactions may stabilize MR binding to DNA (Fig. 5C). One possibility is that the intercomplex interactions reduce disassociations of MRN from DNA. Alternatively, the intercomplex interactions at the apex of the coiled-coil domains of Rad50 through Zn2+-coordinated binding induces conformation change of the globular domain of MR bound to DNA, causing increased affinity of MR for DNA interaction. It was described that binding of MRN globular domain to DNA causes conformational change of the coiled-coil arms of Rad50, which favors the intercomplex association of MRN through the zinc hook motif by the coordination of a Zn2+ ion (11). Our studies further support a cross-talk between the distal Rad50 zinc hook motif and the MR globular DNA-binding domain, whereby the Rad50 hook interaction may induce conformational change of MR globular domain, thereby affecting MR DNA binding.
We showed that Zn2+ induces more stable association of wild type MR but not the MR(HK-NA) mutant with DNA in vitro, suggesting that Rad50 zinc hook domain-dependent MRN intercomplex interactions may lead to more stable DNA binding and thus contribute to more stable initial binding of MR to chromosomal DSBs. However, when DSBs are generated in the context of chromatin, the regulation is complex, and other mechanisms may also be involved for the Rad50 zinc hook domain to promote the initial binding of MRN to chromosomal DSBs. For instance, the MRN intercomplex associations through the Rad50 zinc hook domain may cause conformational change of MRN and create new docking sites for other proteins to associate with MRN and strengthen the interaction of MRN with chromosomal DSBs in vivo.
Using Xenopus extracts, it was described that ATP-dependent transaction activities of Rad50 are important for DNA tethering function in vitro, which increases the local concentration of damaged DNA to facilitate ATM activation (38). The Rad50 zinc hook domain tethers DNA in the in vitro analysis (9–11), but severe defects of the Rad50 hook mutants in being recruited to chromosomal DSBs would also abolish the MRN tethering activity in vivo by loss of chromosomal binding. Thus, impaired DSB recruitment and chromosomal DSB binding activity in the Rad50 hook mutants are the major causes for defective ATM activation to initiate DNA damage responses.
In yeast, the Mre11 complex is required for holding the two broken chromosomes together during DSB repair (12, 13); however, such chromosomal end tethering function in mammalian cells appears to be more dependent on the Ku complex rather than MRN (14). Instead, we demonstrate that the Rad50 hook mutants are impaired in the recruitment of MRN to chromosomal DSBs in mammalian cells, leading to severe defects in ATM checkpoint activation, DSB end resection, HR-mediated DSB repair, and activation of the ATR checkpoint pathway. We propose that the loss of chromosomal DSB recruitment is the major cause for the severe defects observed in the Rad50 hook mutants in mammalian cells. MRN is a well conserved DNA repair protein complex, and the Rad50 hook mutants in yeast also exhibit severe impairment in various DNA damage responses (8). It remains to be determined whether this newly identified function of the Rad50 zinc hook domain in the recruitment of MRN to chromosomal DSBs is conserved in yeast and in other organisms. These studies will help understand the underlying mechanisms of how MRN participates in DSB repair, contributing to DNA damage responses and maintenance of genome stability.
Acknowledgments
We thank members of the Wu laboratory for helpful discussions. We also thank Dr. Tanya T. Paull (University of Texas at Austin) for sharing Mre11 and Nbs1 baculovirus constructs and Dr. John Petrini (Memorial Sloan-Kettering Cancer Center) for providing Rad50 cDNA construct.
This work was supported, in whole or in part, by National Institutes of Health Grants CA102361, GM080677, CA140972, and CA102361-07S1 (to X. W). This work was also supported by funds from the Beckman Laser Institute Inc., Foundation (to M. W. B.).

This article contains supplemental Figs. S1–S5.
- MRN
- Mre11-Rad50-Nbs1
- DSB
- double-stranded break
- HR
- homologous recombination
- dsDNA
- double-stranded DNA
- EBFP
- enhanced blue fluorescence protein
- Gy
- gray.
REFERENCES
- 1. Stracker T. H., Petrini J. H. (2011) The MRE11 complex. Starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. D'Amours D., Jackson S. P. (2002) The Mre11 complex. At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3, 317–327 [DOI] [PubMed] [Google Scholar]
- 3. Paull T. T., Lee J. H. (2005) The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 4, 737–740 [DOI] [PubMed] [Google Scholar]
- 4. Takeda S., Nakamura K., Taniguchi Y., Paull T. T. (2007) Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol. Cell 28, 351–352 [DOI] [PubMed] [Google Scholar]
- 5. Hopfner K. P., Karcher A., Craig L., Woo T. T., Carney J. P., Tainer J. A. (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105, 473–485 [DOI] [PubMed] [Google Scholar]
- 6. Williams R. S., Williams J. S., Tainer J. A. (2007) Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520 [DOI] [PubMed] [Google Scholar]
- 7. Hopfner K. P., Craig L., Moncalian G., Zinkel R. A., Usui T., Owen B. A., Karcher A., Henderson B., Bodmer J. L., McMurray C. T., Carney J. P., Petrini J. H., Tainer J. A. (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418, 562–566 [DOI] [PubMed] [Google Scholar]
- 8. Wiltzius J. J., Hohl M., Fleming J. C., Petrini J. H. (2005) The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12, 403–407 [DOI] [PubMed] [Google Scholar]
- 9. Costanzo V., Paull T., Gottesman M., Gautier J. (2004) Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2, E110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Jager M., van Noort J., van Gent D. C., Dekker C., Kanaar R., Wyman C. (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135 [DOI] [PubMed] [Google Scholar]
- 11. Moreno-Herrero F., de Jager M., Dekker N. H., Kanaar R., Wyman C., Dekker C. (2005) Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437, 440–443 [DOI] [PubMed] [Google Scholar]
- 12. Kaye J. A., Melo J. A., Cheung S. K., Vaze M. B., Haber J. E., Toczyski D. P. (2004) DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14, 2096–2106 [DOI] [PubMed] [Google Scholar]
- 13. Lobachev K., Vitriol E., Stemple J., Resnick M. A., Bloom K. (2004) Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14, 2107–2112 [DOI] [PubMed] [Google Scholar]
- 14. Soutoglou E., Dorn J. F., Sengupta K., Jasin M., Nussenzweig A., Ried T., Danuser G., Misteli T. (2007) Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masutomi K., Yu E. Y., Khurts S., Ben-Porath I., Currier J. L., Metz G. B., Brooks M. W., Kaneko S., Murakami S., DeCaprio J. A., Weinberg R. A., Stewart S. A., Hahn W. C. (2003) Telomerase maintains telomere structure in normal human cells. Cell 114, 241–253 [DOI] [PubMed] [Google Scholar]
- 16. Ai H. W., Shaner N. C., Cheng Z., Tsien R. Y., Campbell R. E. (2007) Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46, 5904–5910 [DOI] [PubMed] [Google Scholar]
- 17. Paull T. T., Gellert M. (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 [DOI] [PubMed] [Google Scholar]
- 18. Wu X., Ranganathan V., Weisman D. S., Heine W. F., Ciccone D. N., O'Neill T. B., Crick K. E., Pierce K. A., Lane W. S., Rathbun G., Livingston D. M., Weaver D. T. (2000) ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405, 477–482 [DOI] [PubMed] [Google Scholar]
- 19. Wu X., Avni D., Chiba T., Yan F., Zhao Q., Lin Y., Heng H., Livingston D. (2004) SV40 T antigen interacts with Nbs1 to disrupt DNA replication control. Genes Dev. 18, 1305–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Nievera C. J., Lee A. Y., Wu X. (2008) Cell cycle-dependent complex formation of BRCA1·CtIP·MRN is important for DNA double-strand break repair. J. Biol. Chem. 283, 7713–7720 [DOI] [PubMed] [Google Scholar]
- 21. Olson E., Nievera C. J., Liu E., Lee A. Y., Chen L., Wu X. (2007) The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell Biol. 27, 6053–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Botvinick E. L., Berns M. W. (2005) Internet-based robotic laser scissors and tweezers microscopy. Microsc. Res. Tech. 68, 65–74 [DOI] [PubMed] [Google Scholar]
- 23. Liu E., Lee A. Y., Chiba T., Olson E., Sun P., Wu X. (2007) The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J. Cell Biol. 179, 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H., Shao Z., Shi L. Z., Hwang P. Y., Truong L. N., Berns M. W., Chen D. J., Wu X. (2012) CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 287, 21471–21480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilson I. A., Haft D. H., Getzoff E. D., Tainer J. A., Lerner R. A., Brenner S. (1985) Identical short peptide sequences in unrelated proteins can have different conformations. A testing ground for theories of immune recognition. Proc. Natl. Acad. Sci. U.S.A. 82, 5255–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lai J. S., Herr W. (1992) Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U.S.A. 89, 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mirzoeva O. K., Petrini J. H. (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell Biol. 21, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lukas C., Falck J., Bartkova J., Bartek J., Lukas J. (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5, 255–260 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. H., Paull T. T. (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26, 7741–7748 [DOI] [PubMed] [Google Scholar]
- 30. Chapman J. R., Jackson S. P. (2008) Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 9, 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu L., Luo K., Lou Z., Chen J. (2008) MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 105, 11200–11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J. (2008) Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell Biol. 181, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spycher C., Miller E. S., Townsend K., Pavic L., Morrice N. A., Janscak P., Stewart G. S., Stucki M. (2008) Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell Biol. 181, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Celeste A., Fernandez-Capetillo O., Kruhlak M. J., Pilch D. R., Staudt D. W., Lee A., Bonner R. F., Bonner W. M., Nussenzweig A. (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5, 675–679 [DOI] [PubMed] [Google Scholar]
- 35. Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S. P., Bartek J., Lukas J. (2004) Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8, 37–45 [DOI] [PubMed] [Google Scholar]
- 37. Hohl M., Kwon Y., Galván S. M., Xue X., Tous C., Aguilera A., Sung P., Petrini J. H. (2011) The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 18, 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dupré A., Boyer-Chatenet L., Gautier J. (2006) Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 13, 451–457 [DOI] [PubMed] [Google Scholar]