This study shows that the vegetative class of plant actins and the cytoplasmic class of animal actins share conserved functions that were inherited from an ancestral protist actin sequence. Thus, some single-celled protists contain actin that can perform the complex processes required for multicellular development.
Abstract
Actin is an essential multifunctional protein encoded by two distinct ancient classes of genes in animals (cytoplasmic and muscle) and plants (vegetative and reproductive). The prevailing view is that each class of actin variants is functionally distinct. However, we propose that the vegetative plant and cytoplasmic animal variants have conserved functional competence for spatial development inherited from an ancestral protist actin sequence. To test this idea, we ectopically expressed animal and protist actins in Arabidopsis thaliana double vegetative actin mutants that are dramatically altered in cell and organ morphologies. We found that expression of cytoplasmic actins from humans and even a highly divergent invertebrate Ciona intestinalis qualitatively and quantitatively suppressed the root cell polarity and organ defects of act8 act7 mutants and moderately suppressed the root-hairless phenotype of act2 act8 mutants. By contrast, human muscle actins were unable to support prominently any aspect of plant development. Furthermore, actins from three protists representing Choanozoa, Archamoeba, and green algae efficiently suppressed all the phenotypes of both the plant mutants. Remarkably, these data imply that actin’s competence to carry out a complex suite of processes essential for multicellular development was already fully developed in single-celled protists and evolved nonprogressively from protists to plants and animals.
INTRODUCTION
Actin is a highly conserved ubiquitous protein that is essential for several important cellular processes, including endocytosis, cell motility, cell division and cytokinesis, vesicle and organelle movement, cell signaling, and the establishment and maintenance of cell junctions, cell shape, and cell polarity (Staiger and Blanchoin, 2006; Pollard and Cooper, 2009). In animals and plants, actin is often encoded as multiple variants that are all vital for normal cell growth and organ development. Modern estimations of the tree of life built from thousands of sequence comparisons indicate that the plant and animal kingdoms shared a common protist ancestor more than 1.5 billion years ago (Figure 1A) and the deuterostome animals and vascular plants evolved independently ∼800 and 600 million years ago, respectively (DeVries et al., 2006; Keeling, 2007; Zimmer et al., 2007). During the evolution from protist to higher eukaryotes, the sequences of actin variants have been relatively conserved, yet plants and animals evolved distinct classes of actin variants, perhaps to support their independent multicellular ontogenies.
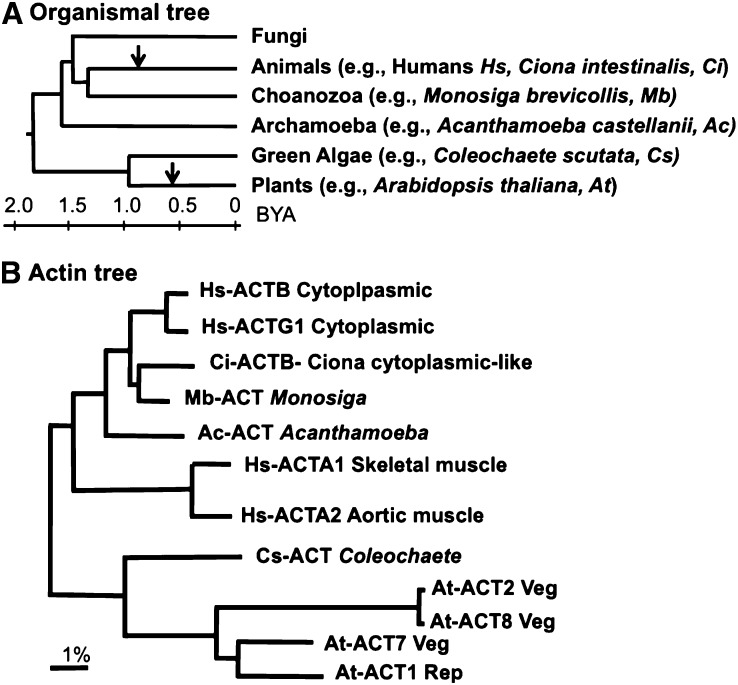
Figure 1.
Phylogenetic Relationships among Various Eukaryotes and Actins.
(A) An organismal tree of life comparing the four eukaryotic kingdoms: Fungi, animals, plants, and protists (Hedges et al., 2004; Keeling, 2007). The approximate divergence times (billions of years ago [BYA]) from a common ancestry are indicated in a scale. The points of origin of deuterostome animals and vascular plants are marked with arrows.
(B) An actin tree comparing the Arabidopsis plant, human, and C. intestinalis animal and protist protein sequences that were examined or discussed in detail in this study. The scale bar represents percent divergence in amino acid sequences.
In both animals and plants, most actin genes are essential for specific aspects of development as evident from genetic studies of various mutants. For example, deficiencies in individual actin genes in mice or Arabidopsis thaliana result in embryo lethality, low viability, and/or severe developmental defects within different organs and cell types (Asmussen et al., 1998; Gilliland et al., 1998; Shawlot et al., 1998; Crawford et al., 2002; Liu et al., 2003; Sonnemann et al., 2006; Kandasamy et al., 2009; Meagher et al., 2011). In Arabidopsis, deficiency in one of the three constitutively expressed actins, ACT7, severely impairs root organ development and results in minor shoot defects, whereas lack of the other two constitutive actins, ACT2 and ACT8, affects root hair cell tip growth (Kandasamy et al., 2009). Missense mutations in human actins are also associated with a variety of gene-specific developmental pathologies, including skeletal and smooth muscle myopathies (Sparrow et al., 2003; Donnell et al., 2008), midline malformations (Procaccio et al., 2006), neutrophil dysfunction (Nunoi et al., 1999), and deafness (Zhu et al., 2003; Bryan et al., 2006).
Based on their sequence phylogeny and expression pattern, the multiple actin variants encoded in plants and animals are each grouped into distinct ancient classes. Accordingly, the cytoplasmic and muscle actin variants in humans or mice form two discrete classes that are 6 to 7% divergent from each other in their amino acid sequences (Figure 1B; see Supplemental Table 1 online). These two classes of animal actins arose early in the deuterostome animal lineage and are readily identified as distinct protein classes in the sea squirt, Ciona intestinalis, which shared a common ancestor with humans ∼700 million years ago. Similarly, the vegetative and reproductive classes of actin variants in vascular plants, such as maize (Zea mays) or Arabidopsis, arose early in the vascular plant lineage, ∼450 million years ago, and are 7 to 8% divergent from each other in protein sequence and 10 to 14% divergent from the deuterostome animal sequences (Figure 1B; see Supplemental Table 1 online). The protein sequences of these four separate classes of actin variants are distinct from each other within their phyla and highly conserved within their lineages. Because the common ancestral actin sequence shared by plants and animals must be from a very ancient single-celled protist (Figure 1A), a long-held view is that the different conserved classes of plant and animal actin variants each have evolved distinct functions that are essential for cell, tissue, and organ development (Ponte et al., 1983; Hightower and Meagher, 1986; Meagher, 1995).
This view that these four classes of actins are all functionally distinct is further supported by the failure of divergent classes of plant and animal actin variants to efficiently suppress actin null mutations in different actins within or across species (Karlsson et al., 1991; Kumar et al., 1997; Fyrberg et al., 1998; Brault et al., 1999; Schildmeyer et al., 2000; Kandasamy et al., 2007). For example, in Drosophila melanogaster, a null mutation in the 88F gene encoding an adult flight muscle actin produces a flightless phenotype, which can be rescued by a wild-type copy of 88F or the other flight muscle actin 79B, but not by the fly larval muscle or cytoplasmic actins or by human cytoplasmic β-actin Hs-ACTB (Fyrberg et al., 1998; Brault et al., 1999). In mice, an αcardiac-actin knockout results in lethality during either embryonic or prenatal development, with profound disorganization of cardiac myofibrils (Kumar et al., 1997). Overexpression of γsmooth-actin only partially rescues the αcardiac-actin knockout, and in those few mice that survive to adulthood, their hearts are weak and enlarged, demonstrating that αcardiac-actin and γsmooth-actin variants make distinct contributions to muscle cell function. Budding yeast actin null mutations are only partially suppressed by chicken β-actin, which supports yeast cell viability, but cause slow growth, aberrant morphology, and temperature-sensitive lethality (Karlsson et al., 1991). By contrast, we found that neither vegetative nor reproductive actins from Arabidopsis could suppress the inviability phenotype caused by the yeast actin null mutation (Kandasamy et al., 2007). In addition, the ectopic expression of Arabidopsis reproductive actin, At-ACT1, but not the overexpression of a vegetative actin, At-ACT2, in Arabidopsis vegetative tissues causes extremely aberrant development of organs and cell types with altered organization of F-actin structures (Kandasamy et al., 2002). These aberrant phenotypes are entirely suppressed by the overexpression of reproductive, but not vegetative, actin binding proteins (Kandasamy et al., 2007). Thus, it is reasonable to propose that the loss of appropriate interactions with actin binding proteins may be at the heart of the functional differences among actin variants. With this preponderance of evidence from genetic suppression studies, the simplest deduction is that the ancient classes of plant and animal actin protein variants are functionally distinct.
However, none of these genetic suppression studies examined the possible conservation of developmental functions among animal cytoplasmic and plant vegetative classes of actins and their ancestral protist actins. Based on experimental data and comparative genomics, the animal cytoplasmic and plant vegetative actins direct numerous related cellular functions that are essential for the spatial development of cells, tissues, and organs. These functions include the cytoplasmic activities of maintaining cell polarity, movement of all organelles in the cytoplasm, such as trafficking of vesicles for exo- and endocytosis and the positioning of membrane receptors, and the nuclear activities of moving various RNA polymerases, trafficking nuclear structures and macromolecules, and remodeling chromatin. Taken together, all such activities may comprise actin’s ability to form dynamic three-dimensional cellular structures and maintain them over various time periods to influence development, which we define here as actin’s competence for spatial development. Comparative genomics data and the morphology of extant protists (e.g., Choanozoa, Archamoeba, and green algae; Figure 1A) suggest that ancestral protists closely related to animals and/or plants also contain similar sophisticated actin systems capable of supporting many of these cytoplasmic and nuclear processes (Anderson et al., 2005; Derelle et al., 2006; Merchant et al., 2007; King et al., 2008; Fritz-Laylin et al., 2010). Consequently, we considered an alternative hypothesis that cytoplasmic animal and vegetative plant actins inherited their competence for spatial development from a common ancestral protist actin. We provide strong initial support for this hypothesis by demonstrating that a subset of divergent actins from animals and protists (Figure 1B; see Supplemental Data Set 1 online), which have not shared common ancestry with plant actins for more than a billion years, efficiently suppressed a diverse suite of cell and organ developmental defects of Arabidopsis mutants deficient in vegetative actins, while more specialized muscle actins failed to support markedly any plant developmental functions.
RESULTS
Plant Vegetative Actin Double Mutants Exhibit Distinct Developmental Defects
Arabidopsis encodes eight actin variants, three vegetative and five reproductive. Among the three vegetative actins, ACT7 is expressed strongly in young seedlings and developing organs and regulated by plant hormones, whereas ACT2 is constitutively abundant in adult plants and mature organs (Meagher et al., 1999; Kandasamy et al., 2001, 2009; Gilliland et al., 2003). These two variants are more prevalent in all plant organs than ACT8, which is the most weakly expressed actin in both young and mature organs. In shoots, 10 to 15% of the total actin is ACT8 compared with 45 to 50% ACT2 and 40 to 45% ACT7 (Meagher et al., 1999; Kandasamy et al., 2009). These variants exhibit functional specificity such that the lack of ACT7 mainly affects cell polarity (Gilliland et al., 2003) and organ growth as shown for roots in Figures 2A to 2C, while the lack of ACT2 (Gilliland et al., 2002) or ACT8 (Figure 2A) affects only root hair growth. Due to partial functional redundancy, the double homozygous null mutants often exhibit stronger and more distinct developmental phenotypes than the single mutants. For example, double mutants lacking in ACT8 and ACT7 (act8-2 act7-4; Kandasamy et al., 2009) are highly dwarfed with strong root and shoot growth phenotypes (Figure 2), but they are able to survive and set seeds. Double mutants lacking in ACT2 and ACT8 (act2-1 act8-2) develop into relatively normal plants, but with bald roots having no root hairs (Kandasamy et al., 2009). The trichoblast cells of act2-1 act8-2 roots show no tip growth from the root hair bulges (i.e., no cell elongation). On the other hand, double mutants null for ACT2 and ACT7 (e.g., act2-1 act7-4) are extremely dwarfed and nearly infertile and are difficult to manipulate in the laboratory (Gilliland et al., 2002; Kandasamy et al., 2009). Therefore, the act8-2 act7-4 and act2-1 act8-2 double mutants are ideal for genetic suppression studies to examine the specific roles of diverse actins on shoot and root organ development and root hair cell tip growth, respectively. The morphological and cellular phenotypes of act2-1 act7-4 and act2-1 act8-2 double mutant plants were fully characterized previously (Kandasamy et al., 2009); hence, only the defects of the act8-2 act7-4 plants that are used throughout this article are presented in Figures 2A to 2O and described here in detail.
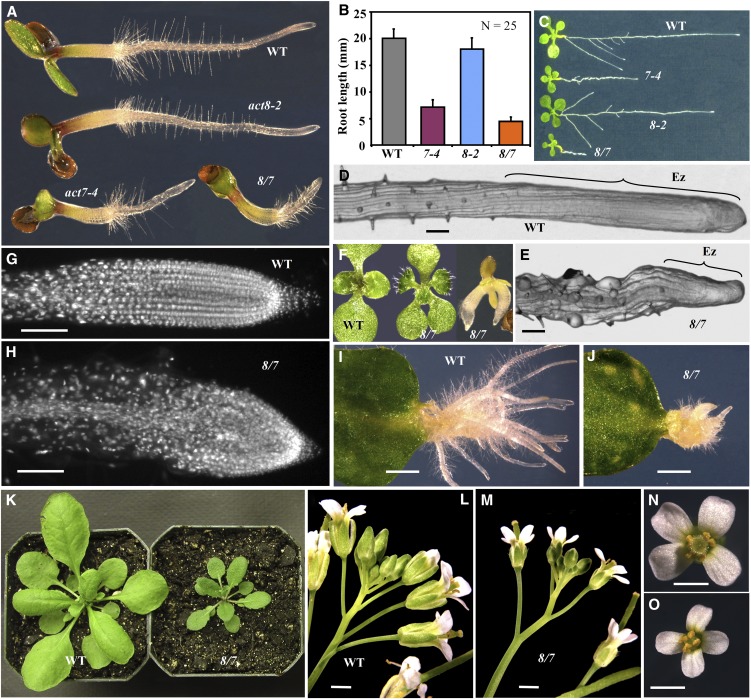
Figure 2.
Phenotypic Characterization of the act8-2 act7-4 Double Mutant.
(A) to (C) Comparison of root growth of the act8-2 (8-2) and act7-4 (7-4) single mutants and the act8-2 act7-4 (8/7) double mutant and the wild type (WT).
(A) Three-day-old seedlings. Note the shorter root hairs in the act8-2 seedling than the wild type.
(B) Root length of 3-d-old seedlings. The error bars represent sd.
(C) Eight-day-old seedlings.
(D) and (E) Root tips. Ez, elongation zone.
(F) Aberrant mutant seedlings with three cotyledons (middle) or fused embryos with multiple radicals (right). Wild-type seedling with two cotyledons is shown on the left.
(G) and (H) DAPI-stained root tips showing loss of cell polarity in the 8/7 double mutant relative to the wild type.
(I) and (J) Adventitious root growth from leaves.
(K) Twenty-six-day-old plants.
(L) and (M) Inflorescences.
(N) and (O) Flowers.
Bars = 50 μm (D), (E), (G), and (H), 2 mm in (I) and (J), and 1 mm in (L) to (O).
The act8-2 act7-4 double mutant seedlings had dwarf roots that were <25% the length of the wild type (Figures 2A to 2C). The double mutant root phenotype was significantly more severe than in any of the single mutants such as the ACT7 mutant (act7-4) that had roots 35 to 40% the length of wild type or the ACT8 mutant (act8-2) that had root length similar to wild type. The root growth defects in the double mutants became more pronounced as the plants grew, severely affecting the whole root architecture (Figure 2C). The act8-2 act7-4 roots had an elongation zone that was almost three times smaller (the region from the root tip to the first root hair) than the wild type and had a rough surface with irregularly bulged epidermal cells compared with the smooth surface of wild-type root apices (Figures 2D and 2E). Approximately 8% of act8-2 act7-4 seeds contained multiple, often fused, embryos or aberrant embryos that upon germination produced seedlings with three or four cotyledons instead of the usual two, a phenotype seldom observed in the wild type (Figure 2F). Because the act8-2 act7-4 plants had defects in the growth and elongation of primary roots, we were interested in determining if these plants were also deficient in adventitious root initiation and growth. Therefore, we induced roots from leaves by incubating them with the phytohormone auxin. Following auxin treatment, the act8-2 act7-4 mutant leaves produced a few root primordia, but they were unable to grow into long adventitious roots as in the wild type (Figures 2I and 2J). These results demonstrate that the act8-2 act7-4 plants were defective in the growth of both primary and adventitious roots.
To understand the possible mechanism behind the retarded root growth of act8-2 act7-4 plants, we examined the cellular organization of root tips. The act8-2 act7-4 mutant root tips had irregular masses of cells as a result of oblique cell wall formation during cell division with an apparent disruption in cell polarity (Figure 2H), as shown previously for the act7-4 single mutant (Gilliland et al., 2003). In the wild-type root, on the other hand, the elongation zone contained regular files of cells as a result of transversely aligned planes of cell division (Figure 2G). In addition to the obvious root development phenotypes, the act8-2 act7-4 plants revealed dwarfed shoot (Figure 2K) and inflorescence (Figures 2L and 2M) phenotypes with distinctly smaller rosette leaves (40% narrower than the wild type; Figure 2K) and flowers (30% smaller than the wild type; Figures 2N and 2O).
Human Cytoplasmic β-Actin ACTB, but Not α-Skeletal Muscle Actin ACTA1, Restores All the Developmental Defects of act8-2 act7-4 Plants
To determine whether deuterostome actin variants could substitute for plant vegetative actin functions, we expressed human cytoplasmic β-actin ACTB and skeletal muscle α1-actin ACTA1 in the act8-2 act7-4 mutant background under control of the constitutively active At-ACT2 regulatory sequences (A2p) (see Methods). We screened more than 15 independent transformants each expressing the A2p:HsACTB and A2p:HsACTA1 constructs for the suppression of mutant phenotypes. For that screening, seven distinct developmental parameters were examined: root length of seedlings, root architecture of young plants, cellular organization at the root apex, adventitious root development from leaf explants, organization of embryos in developing seeds, plant/leaf morphology, and inflorescence/flower architecture (Figures 3A to 3O, 4A, 4B, 5A, and 5B).
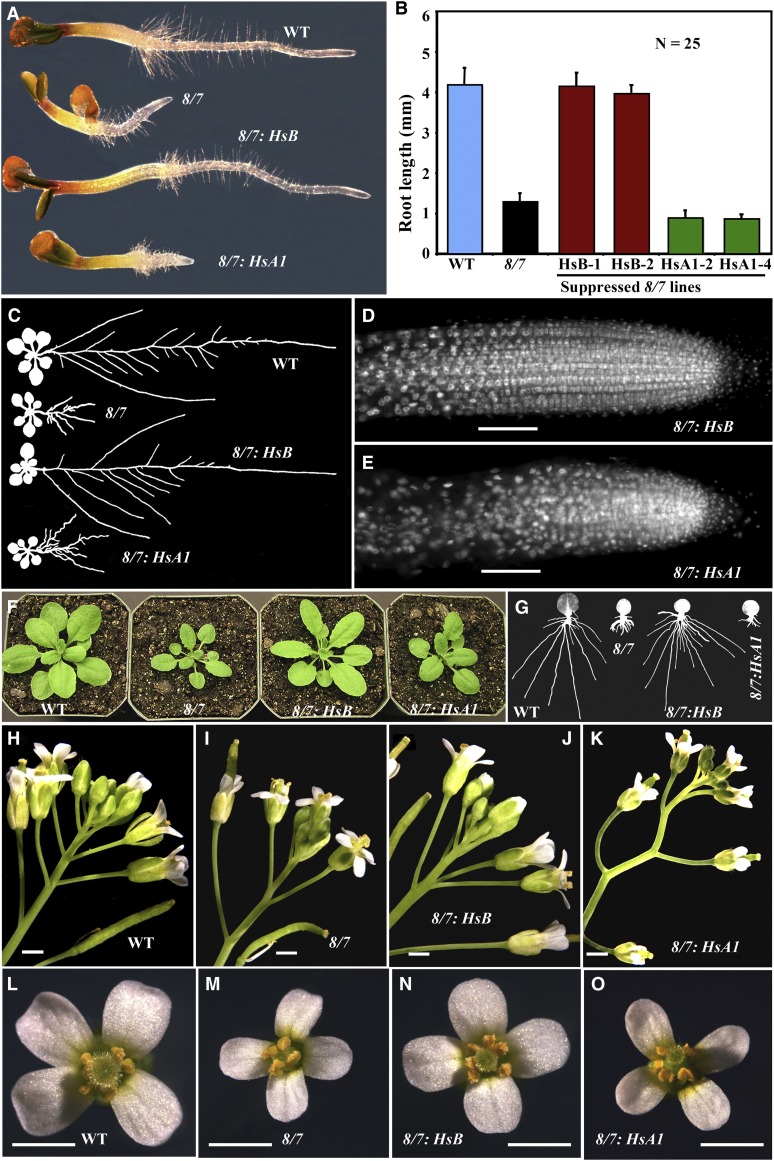
Figure 3.
Rescue of act8-2 act7-4 Double Mutant Phenotypes by Human Cytoplasmic ACTB and Skeletal Muscle ACTA1 Actin Variants.
(A) Morphology of 3-d-old seedlings comparing the wild type (WT), act8-2 act7-4 (8/7) mutant, and the mutant expressing human ACTB (HsB) or ACTA1 (HsA1).
(B) Root lengths of 3-d-old seedlings. Two independent transformed lines are shown for each construct.
(C) Root architecture of 14-d-old plants.
(D) and (E) DAPI DNA-stained root tips showing cellular organization (cf. to Figures 2G and 2H).
(F) Twenty-five-day-old plants.
(G) Adventitious root growth from leaves.
(H) to (K) Inflorescences.
(L) to (O) Flowers.
Bars = 50 μm in (D) and (E) and 1 mm in (H) to (O).
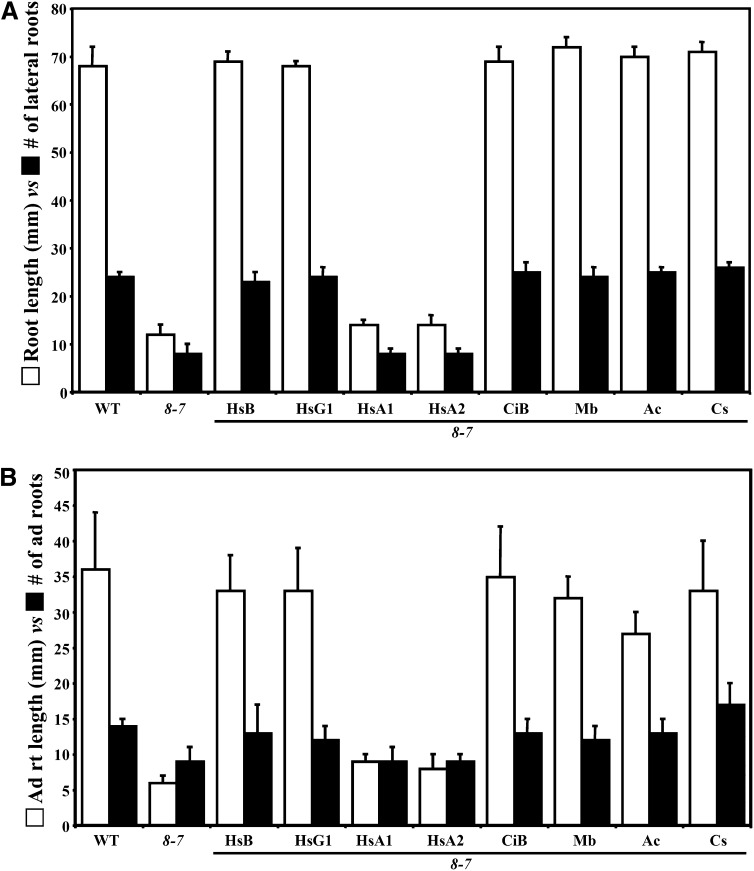
Figure 4.
Suppression of Root Architecture and Adventitious Root Growth Phenotypes by Diverse Actins.
(A) Root length (white, left bar in each pair) versus number of lateral roots (black, right bar in each pair) produced from 14-d-old seedlings of wild-type (WT), act8-2 act7-4 double mutant (8/7), and various transgenic lines expressing diverse actins.
(B) Adventitious root length (white) versus number of roots (black) produced from the leaf explants after 4 d of auxin treatment followed by 6 d of growth on MS medium. The data represent average values from three biological replicates with each experiment containing 10 samples for each line shown. The error bars represent sd. HsB, Hs-ACTB; HsG1, Hs-ACTG1; HsA1, Hs-ACTA1; HsA2, Hs-ACTA2 (human); CiB, Ci-ACTB1 (C. intestinalis); Mb, Mb-ACT (M. brevicollis); Ac, Ac-ACT (A. castellanii); Cs, Cs-ACT (C. scutata).
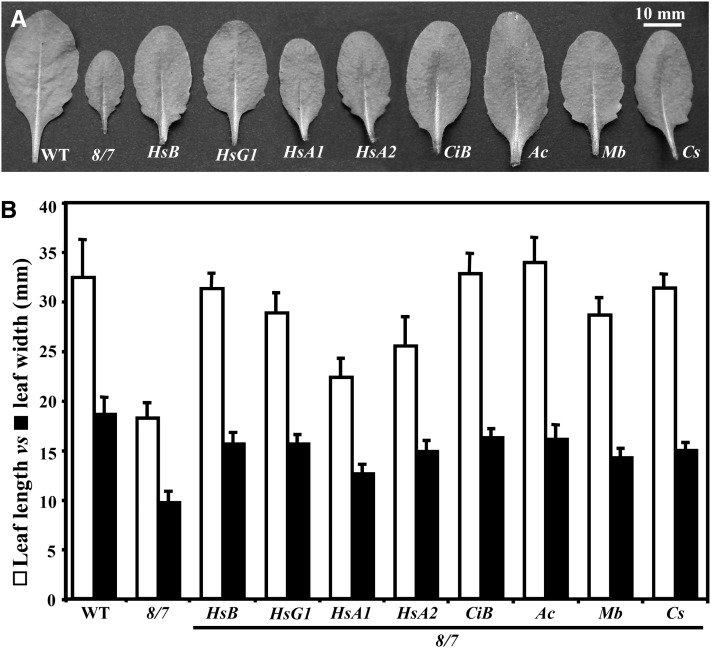
Figure 5.
Suppression of the Leaf Morphology Phenotype by Diverse Actins.
(A) The largest rosette leaf from 25-d-old plants. WT, the wild type.
(B) Leaf length (white) versus leaf width (black) of wild-type, act8-2 act7-4 double mutant (8/7), and various transgenic lines expressing diverse actins. Hs, human; Ci, C. intestinalis; Mb, M. brevicollis; Ac, A. castellanii; and Cs, C. scutata protist actins. The data correspond to average values from the two largest leaf samples from 10 plants (24 d) for each line shown, and the error bars represent sd.
The human cytoplasmic β-actin variant ACTB fully rescued the root morphology defects of 3-d-old act8-2 act7-4 seedlings (Figure 3A); hence, the transformed seedlings had root lengths similar to the wild type (Figure 3B). The mature roots of 12- to 14-d-old act8-2 act7-4/A2p:HsACTB plants had normal root architecture with the length of the primary roots and the number of lateral roots equivalent to the wild type (Figures 3C and 4A). Hence, lateral root primordia were initiated at the appropriate intervals. The root tips of the suppressed plants had cellular organization and cell polarity comparable to the wild type with cells in regular rows (Figures 2G and 3D). Human cytoplasmic β-actin also restored normal morphology and size to shoots (Figure 3F), leaves (Figures 5A and 5B), inflorescences (Figures 3H to 3J), and flowers (Figures 3L to 3N). Moreover, we found the application of auxin induced adventitious roots from leaves of β-actin suppression lines with an efficiency similar to that of the wild type (Figures 3G and 4B). Almost 100% of transgenic seeds also contained normal embryos with the usual two cotyledons and one radical. We obtained similar results for all 15 independent transgenic Hs-ACTB–expressing plant lines examined. On the other hand, expression of human α-skeletal muscle actin transgene A2p:HsACTA1 in act8-2 act7-4 plants failed to rescue quantitatively any of the seven developmental phenotypes we examined, and the transformed plants resembled untransformed mutant plants in nearly all aspects (Figures 3 and 4). None of the 15 independent transgenic Hs-ACT1 suppression lines we generated showed restoration of any normal phenotypes, except for the shoots, especially the size of leaves, which appeared somewhat improved in most lines compared with the double mutant (Figures 3F, 5A, and 5B). Mutant suppression data are summarized in Table 1.
Table 1. Summary of Suppression of the Arabidopsis Vegetative Actin Double Mutant Phenotypes by Diverse Actins.
| Actin Transgene in Double Mutant | Root Hair Length (µm)a | Root Length (mm)b,c | Cell Polarityb | Embryo Developmentb | Root Architecture (mm)b,d | Inflorescence Architectureb | Adventitious Root Developmentb,e | Plant Morphologyb,f |
|---|---|---|---|---|---|---|---|---|
| Hs-ACTA1 | 50 ± 8 | 1.2 ± 0.05 | Aberrant | Aberrant | Dwarf (14 ± 1/8 ± 1) | Aberrant | Poor (9 ± 1/9 ± 2) | Semidwarf |
| Hs-ACTA2 | 40 ± 5 | 1.2 ± 0.04 | Aberrant | Aberrant | Dwarf (14 ± 2/8 ± 1) | Aberrant | Poor (8 ± 2/9 ± 1) | Semidwarf |
| Hs-ACTB | 283 ± 23 | 4.4 ± 0.1 | Normal | Normal | Normal (69 ± 2/23 ± 2) | Normal | Robust (33 ± 5/13 ± 4) | Normal |
| Hs-ACTG1 | 238 ± 21 | 4.3 ± 0.1 | Normal | Normal | Normal (68 ± 1/24 ± 2) | Normal | Robust (33 ± 6/12 ± 2) | Normal |
| Ci-ACTB1 | 125 ± 4 | 4.1 ± 0.05 | Normal | Normal | Normal (69 ± 3/25 ± 2) | Normal | Robust (35 ± 7/13 ± 2) | Normal |
| Mb-ACT | 572 ± 6 | 4.4 ± 0.2 | Normal | Normal | Normal (72 ± 2/24 ± 2) | Normal | Robust (32 ± 3/12 ± 2) | Normal |
| Ac-ACT | 478 ± 30 | 4.7 ± 0.2 | Normal | Normal | Normal (70 ± 2/25 ± 1) | Normal | Robust (27 ± 3/13 ± 2) | Normal |
| Cs-ACT | 523 ± 26 | 4.8 ± 0.1 | Normal | Normal | Normal (71 ± 2/26 ± 1) | Normal | Robust (33 ± 7/17 ± 3) | Normal |
| Wild type | 575 ± 29 | 4.4 ± 0.1 | Normal | Normal | Normal (68 ± 4/24 ± 1) | Normal | Robust (36 ± 8/14 ± 1) | Normal |
| 2/8 | 0 | 4.4 ± 0.2 | Normal | Normal | Normal (69 ± 3/26 ± 4) | Normal | Robust (35 ± 5/16 ± 4) | Normal |
| 8/7 | 297 ± 61 | 1.1 ± 0.1 | Aberrant | Aberrant | Dwarf (12 ± 2/8 ± 2) | Aberrant | Poor (6 ± 1/9 ± 2) | Dwarf |
act2-1 8-2 double mutant background.
act8-2 7-4 double mutant background.
Three-day-old seedling.
Fourteen-day-old plant root length versus number of lateral roots.
Length of adventitious roots produced from leaf explants versus number of roots produced.
See Figure 5 for quantification of leaf length versus leaf width. At least 10 independent transformed lines for each transgene revealed this phenotype.
Immunoblot analysis with the monoclonal antibody MAbGEa, which detects both human cytoplasmic Hs-ACTB and muscle Hs-ACTA1 actins (Figure 6A), revealed that all A2p:HsACTB and A2p:HsACTA1 transformed mutant plants examined expressed high levels of human cytoplasmic β-actin or skeletal muscle α1-actin, respectively. The levels of cytoplasmic β-actin or muscle α1-actin in the transgenic plants ranged from being equivalent to wild-type plant actin levels to up to threefold above wild-type levels. Actin expression for two such lines for each construct is shown in Figure 6B. The two A2p:HsACTB lines shown, HsB-1 and HsB-2, expressed almost 1.5 times as much human β-actin compared with wild-type total plant actin levels, and both showed almost full suppression of all phenotypes (Figure 3B). The A2p:HsACTA1–expressing lines HsA1-2 and HsA1-4 produced levels of α1-actin similar to the amount of β-actin produced by the A2p:HsACTB lines, but they were still unable to rescue the mutant phenotype (Figure 3B). We found that plants showing high levels of expression of either human actin variant, approximately threefold above wild type levels, had slightly diminished shoot architecture relative to the wild type, but had normal root growth, similar to the results we observed with strong Arabidopsis ACT2 overexpression (Kandasamy et al., 2002). Because the antibody used in these assays, MAbGEa (Kandasamy et al., 1999), reacts almost equally with the human cytoplasmic β-actin ACTB and skeletal muscle α1-actin ACTA1 (Figure 6A), the failure of phenotypic suppression by Hs-ACTA1 and successful rescue by Hs-ACTB are not due to differences in steady state protein levels.
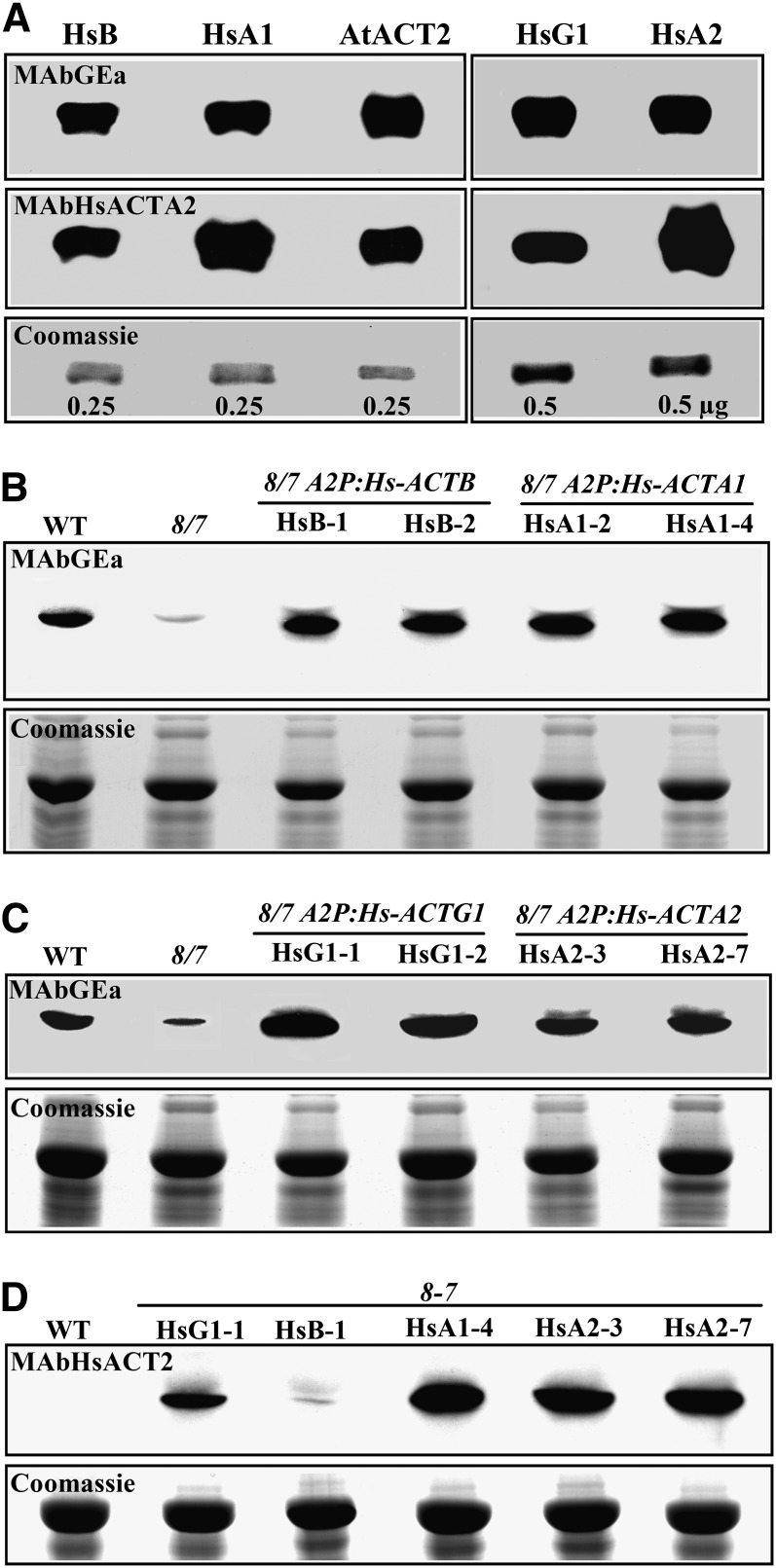
Figure 6.
Immunoblot Analysis of Human Cytoplasmic (Hs-ACTB and Hs-ACTG1) and Muscle Actin (Hs-ACTA1 and Hs-ACTA2) Expression in Transgenic Plants.
(A) Reactivity of antiactin antibodies MAbGEa (top panel) and MAbHsACTA2 (middle panel) with human (HsB and HsA1, native; HsG1 and HsA2, recombinant) and plant (At-ACT2, recombinant) actins. The numbers on the Coomassie blue–stained gel shown in the bottom panel indicate the amount of actin protein loaded.
(B) Expression of Hs-ACTB (cytoplasmic β-actin) and Hs-ACTA1 (muscle α1-actin) in act8 act7 (8/7) A2p:Hs-ACTB and A2p:Hs-ACTA1 plants, respectively, versus total plant actin in 8/7 plants. WT, the wild type.
(C) Expression of Hs-ACTG1 and Hs-ACTA2 in act8 act7 plants. The top panels in (B) and (C) were probed with MAbGEa. The bottom panels show images of Coomassie blue–stained duplicate gels. Protein expression for two independent transformed lines is shown for each transgene.
(D) Protein blot probed with anti-Hs-ACTA2 antibody MAbHsACTA2. Note this antibody reacts strongly with both human muscle actins and poorly with native plant actins or human cytoplasmic actins expressed in plants. All lanes were loaded with 25 µg of total plant protein.
Human Cytoplasmic ACTG1, but Not Aortic Muscle ACTA2, Suppress the Developmental Defects of Plant Vegetative Actin Double Mutants
Our above studies demonstrate that the cytoplasmic Hs-ACTB is very similar to the plant vegetative actins in supporting various developmental functions. To generalize the functional equivalence of the animal cytoplasmic actins and the lack of equivalence for muscle actins with the plant vegetative actins, we examined the suppression of the act8-2 act7-4 plant mutant phenotypes by the other human cytoplasmic actin, ACTG1, and the most divergent muscle actin, ACTA2 (for aortic muscle α2-actin). Hs-ACTG1 is 1.1% divergent from Hs-ACTB, whereas Hs-ACTA2 is 2.2% divergent from Hs-ACTA1 and 5.6% divergent from Hs-ACTG1 (Figure 1B; see Supplemental Table 1 online). Expression of Hs-ACTG1 fully restored root growth to act8-2 act7-4 seedlings, as the young and mature transgenic plants showed no significant difference in root length and architecture from the wild type (Figure 4A, Table 1; see Supplemental Figure 1A online). In addition, Hs-ACTG1 almost fully suppressed all the other prominent developmental defects in act8-2 act7-4 plants, such as adventitious root development, inflorescence/flower architecture, plant (leaf) morphology, and root cell organization and polarity (Figures 4B, 5A, and 5B, Table 1). Thus, Hs-ACTG1 is as efficient as Hs-ACTB in supporting various aspects of Arabidopsis plant development.
On the other hand, aortic muscle actin Hs-ACTA2, like skeletal muscle Hs-ACTA1, cannot efficiently suppress any vegetative plant actin deficiencies, with the exception of mild suppression of the leaf size phenotype (Figures 4 and 5, Table 1; see Supplemental Figure 1A online). Immunoblot analysis revealed that all 15 transgenic A2p:HsACTA2 plants isolated expressed abundant levels of aortic muscle actin, but none showed significant suppression of mutant phenotypes. MAbGEa, which reacts equally with recombinant Hs-ACTG1 (HsG1, Figure 6A) and Hs-ACTA2 (HsA2, Figure 6A) proteins on immunoblots, showed a slightly weaker reactivity with Hs-ACTA2 than Hs-ACTG1 expressed in plants (Figure 6C). Therefore, we tested the plant expression levels of Hs-ACTA2 and Hs-ACTG1 using an antihuman muscle actin antibody. This MAbHsACT2 antibody, which reacts strongly with recombinant muscle actins (HsA1 and HsA2; Figure 6A) and weakly with cytoplasmic actins (HsB and HsG1; Figure 6A), revealed that the levels of Hs-ACTA2 were similar to Hs-ACTA1 muscle actins expressed in transgenic plants (Figure 6D). Thus, even with sufficient levels of expression, Hs-ACTA2 is unable to suppress nearly every one of the plant actin mutant developmental phenotypes.
Human Cytoplasmic Actins, but Not Muscle Actins, Restore Root Hair Development in the act2-1 act8-2 Mutant
In Arabidopsis, ACT2 and ACT8 are essential for normal elongation of root hairs (Kandasamy et al., 2009), which are one of the fastest growing cell types in plants. We wanted to test if human cytoplasmic β-actin and skeletal muscle α1-actin could substitute for these two vegetative actins and rescue the polar tip growth and bald root hair phenotypes of act2-1 act8-2 double mutant plants. The Hs-ACTB and Hs-ACTA1 variants each partially suppressed the root hair cell tip growth defect (Figures 7A to 7C). However, suppression by Hs-ACTB was relatively efficient, with most plant lines having 50 to 60% of wild-type root hair length, while At-ACTA1–suppressed plants never produced root hairs more than 15% of normal length (Figure 7C, Table 1). Immunoblot analysis showed that both human actin variants were equally well expressed in the act2-1 act8-2 mutant background (Figure 7D). The two lines shown for each construct (A2p:HsACTB and A2p:HsACTA1) contained 2 to 3 times more total actin than the wild type. The act2-1 act8-2 mutant plants have almost the same level of total plant actin as the wild type, because of the induction of endogenous ACT7 expression in this actin mutant background (Kandasamy et al., 2009). The degree to which human cytoplasmic β-actin rescued root hair growth is similar to the results we obtained in suppression studies using overexpression of At-ACT7, which restores root hair length in this double mutant to no more than 65% of wild-type levels (Kandasamy et al., 2009). Although the plants expressing β-actin revealed significantly improved root hair growth, high levels of β-actin (e.g., HsB-1) retarded shoot development, as the plants had smaller rosette leaves than the wild type (Figure 7E).
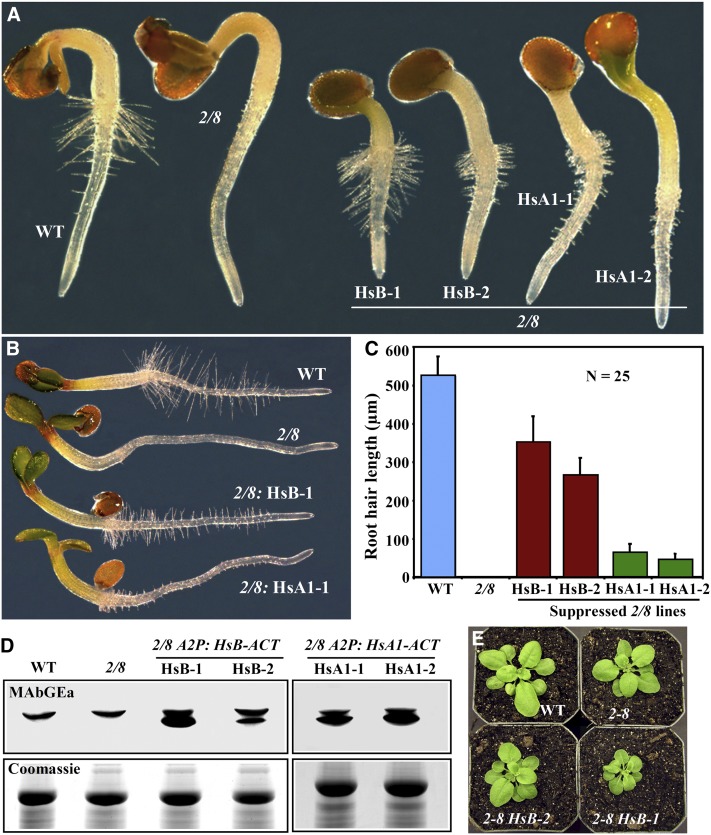
Figure 7.
Rescue of the Root Hair Growth Phenotype of the act2-1 act8-2 Double Mutant by Human ACTB and ACTA1 Actins.
(A) Thirty-six-hour-old seedlings. WT, the wild type.
(B) Three-day-old seedlings.
(C) Root hair length of 3-d-old seedlings. Two independent transformed lines are shown for each construct in (A) and (C). Twenty-five maximally grown root hairs from at least 10 seedlings of each line were used for the measurement. 2/8, act2-1 act8-2; HsB, Hs-ACTB; HsA1, Hs-ACT1. Error bars represent sd.
(D) Immunoblots showing the expression of Hs-ACTB and Hs-ACT1 in two transgenic lines each.
(E) Morphology of 26-d-old act2-1 act8-2 seedlings expressing different levels of Hs-ACTB (see [D]).
Moreover, we tested the effect of expressing Hs-ACTG1 and Hs-ACTA2 on root hair cell elongation in act2-1 act8-2 plants. Similar to Hs-ACTB, Hs-ACTG1 also moderately supported root hair cell elongation (Figure 8; see Supplemental Figure 1B online). The transformed Hs-ACTG1–expressing plants had root hairs that were 40% the length of the wild type, which is ∼10% less suppression than by Hs-ACTB. On the other hand, Hs-ACTA2–induced root hair cell growth to <10% of the wild type (Figure 8, Table 1).
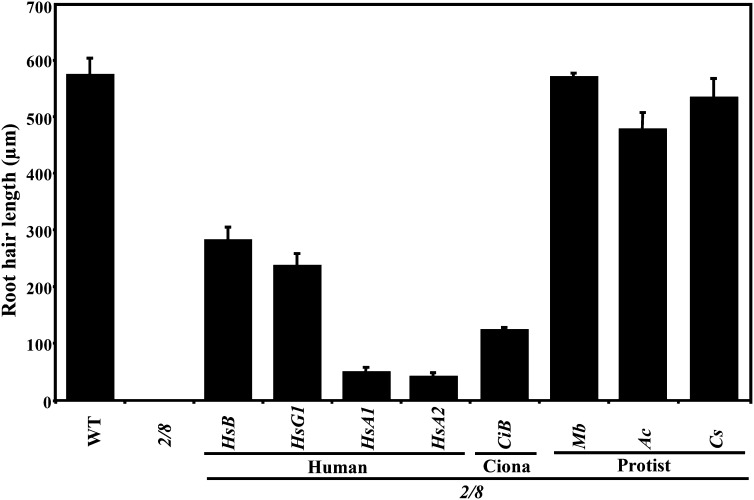
Figure 8.
Suppression of the Root Hair Cell Growth Phenotype by Diverse Actins.
Root hair length of 3-d-old seedlings of wild-type (WT), act2-1 act8-2 double mutant (2/8), and various transgenic lines expressing diverse actins. The data represent average values from three biological replicates, with each experiment containing 25 samples for each line shown. The error bars indicate sd. HsB, Hs-ACTB; HsG1, Hs-ACTG1; HsA1, Hs-ACTA1; HsA2, Hs-ACTA2 (human); CiB, Ci-ACTB1 (C. intestinalis); Mb, Mb-ACT (M. brevicollis); Ac, Ac-ACT (A. castellanii); Cs, Cs-ACT (C. scutata).
Ancestral Ciona β-Actin Also Supports Plant Actin Functions
Arabidopsis and humans have completely independent and divergent ontogenies separated by more than a billion years (Figure 1A); hence, it is extremely unlikely that Arabidopsis vegetative and human cytoplasmic actin variants converged recently on common structures and appropriate protein interactions with the myriad of actin binding proteins that are essential for normal actin cytoskeletal functions. Yet, both these classes of actins functioned similarly in supporting various aspects of plant development. Therefore, we suggest instead that during their evolution from a common ancestral sequence, the animal cytoplasmic and plant vegetative actins preserved ancient functions essential for spatial development. To support this idea, we selected a cytoplasmic-like actin (Ci-ACTB1) from an ancestral deuterostome, C. intestinalis, and tested the suppression of act8-2 act7-4 and act2-1 act8-2 mutant plant phenotypes. The C. intestinalis ACTB1 actin is a clear sequence homolog of human cytoplasmic actins and is 3 to 4% divergent from Hs-ACTB and Hs-ACTG1 (Figure 1B; see Supplemental Table 1 online). C. intestinalis has not shared a common ancestor with humans for 700 million years (DeVries et al., 2006); therefore, the C. intestinalis actin has had ample time to diverge in function and sequence from human actins.
Expression of C. intestinalis cytoplasmic-like actin ACTB1 efficiently suppressed the developmental defects of the act8-2 act7-4 mutant. For example, the root length and architecture of Ci-ACTB1–expressing seedlings was indistinguishable from the wild type or from human ACTB-suppressed plants (Figure 4A; see Supplemental Figure 1A online). Ci-ACTB1 actin protein was also able to fully rescue the adventitious root development (Figure 4B) and leaf morphology (Figure 5). Moreover, the embryo development, inflorescence and flower morphology, and the root apical organization and cell polarity phenotypes of act8-2 act7-4 were also restored to the wild type (Table 1). Immunoblot analysis with MAbGEa, which reacted strongly with recombinant Ci-ACTB1 actin, revealed that Ci-ACTB1 expression levels in all transgenic plant lines were similar to or even higher than total actin levels in wild-type plants (Figures 9A and 9B). However, Ci-ACTB1 expression in act2-1 act8-2 seedlings produced mostly short root hairs that were just 20% the length of the wild type (Figure 8; see Supplemental Figure 1B online). Thus, the cytoplasmic-like actin Ci-ACTB1 fully supports plant growth and development, but only function partially in restoring root hair cell elongation.
Protist Actins Ancestral to Plants and Animals Support Normal Root Hair Tip Growth and Plant Development
To test our hypothesis that the competence for spatial development exhibited by cytoplasmic animal and vegetative plant actins is inherited from a common ancestral protist actin sequence, we examined the ability of protist actins to suppress plant actin deficiency phenotypes. We tested three protist actin sequences that are ancestral to plants and animals: Monosiga brevicollis, Acanthamoeba castellanii, and Coleochaete scutata. M. brevicollis is a single-celled protist belonging to Choanozoa, a phylum that is particularly close to the animal kingdom (King et al., 2008) (Figure 1A). The MbACT actin protein sequence is 2 to 3% divergent from deuterostome cytoplasmic actins, 7 to 8% divergent from muscle actins, and 10 to 13% divergent from plant actins (Figure 1B; see Supplemental Table 1 online). A. castellanii is a single-celled protist that belongs to Archamoeba, a phylum basal to both the fungal and animal kingdoms but more distant from plants. Ac-ACT1 actin is 4 to 14% divergent from the other actins being examined and is also more closely related to cytoplasmic actins than plant actins. C. scutata is a multicellular green algal protist belonging to the Charophytes, the phylum believed to contain the closest living protist relatives to vascular plants but quite distant from animals and fungi. However, the Cs-ACT actin sequence is only slightly closer on the average to plant actin than to animal actin sequences (Figure 1B; see Supplemental Table 1 online).
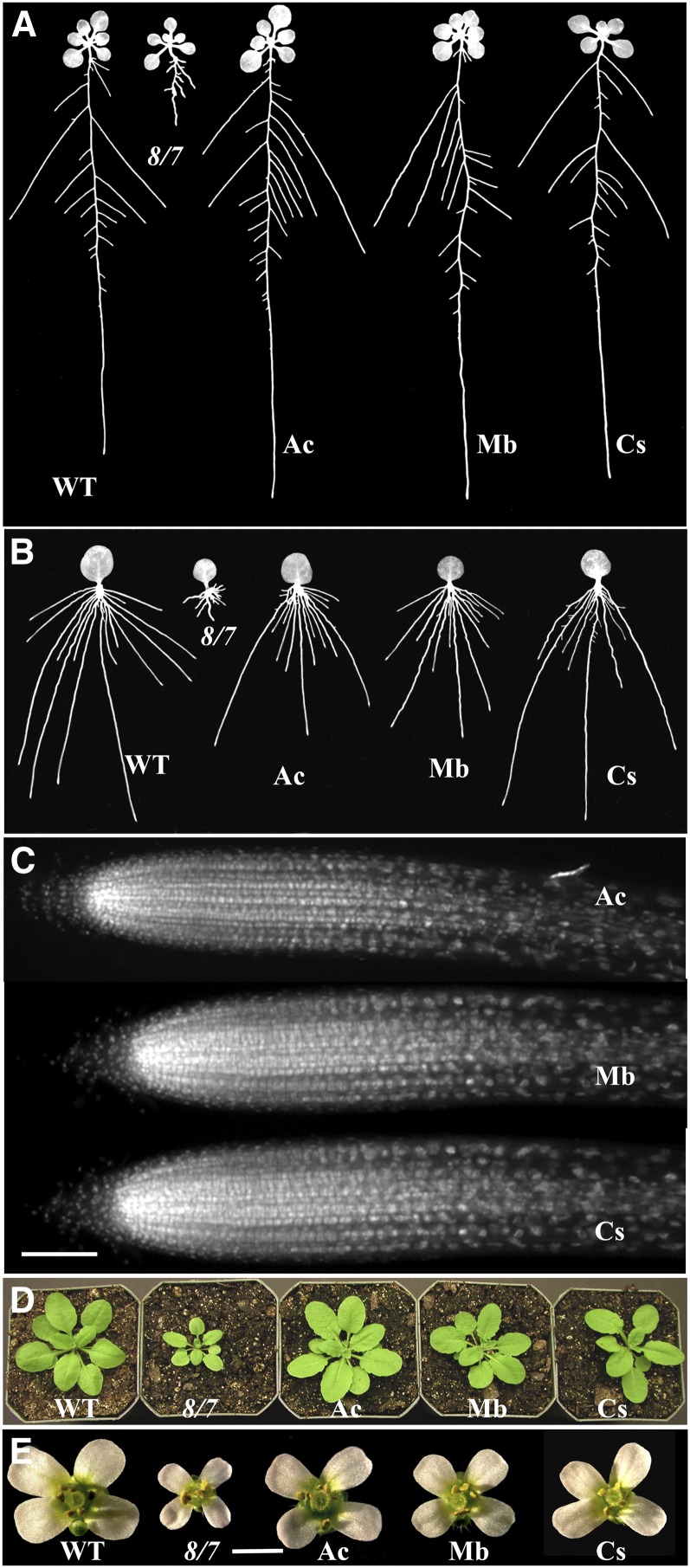
Transgenes expressing the three protist actins A2p:MbACT, A2p:AcACT1, and A2p:CsACT were each effective at suppressing all the developmental defects of the act8-2 act7-4 mutant. For example, seedlings and adult plants containing these transgenes and strongly expressing their encoded proteins (Figure 9C) had normal roots that were of indistinguishable length and architecture from those of wild-type plants (Figures 4A and 10A; see Supplemental Figure 1C online). Other developmental defects of the mutant that were also completely restored to the wild-type phenotype include poor adventitious root development (Figures 4B and 10B), aberrant root apical cell organization and polarity (Figure 10C), dwarf leaf (Figures 5 and 10D) and inflorescence/flower morphology (Figure 10E), and aberrant development of embryos (Table 1). Similarly, expression of Mb-ACT, Ac-ACT1, and Cs-ACT actins efficiently suppressed the root hair cell elongation and tip growth defects of the act2-1 act8-2 mutant, with root hairs being 99, 83, and 90% of wild-type length, respectively (Figure 8; see Supplemental Figure 1D online). Surprisingly, the restoration of root hair elongation by all three protist actins significantly exceeded that by the deuterstome cytoplasmic actins.
Figure 10.
Rescue of act8-2 act7-4 Double Mutant Phenotypes by Protist Actins.
(A) Root architecture of 14-d-old seedlings. WT, the wild type; 8/7, act8-2 act7-4; Ac, A. castellanii; Mb, M. brevicollis; Cs, C. scutata.
(B) Adventitious roots developed from leaves.
(C) DAPI-stained root tips showing cellular organization.
(D) Twenty-six-day-old plants.
(E) Flowers from adult plants.
Bars = 50 μm in (C) and 1 mm in (E).
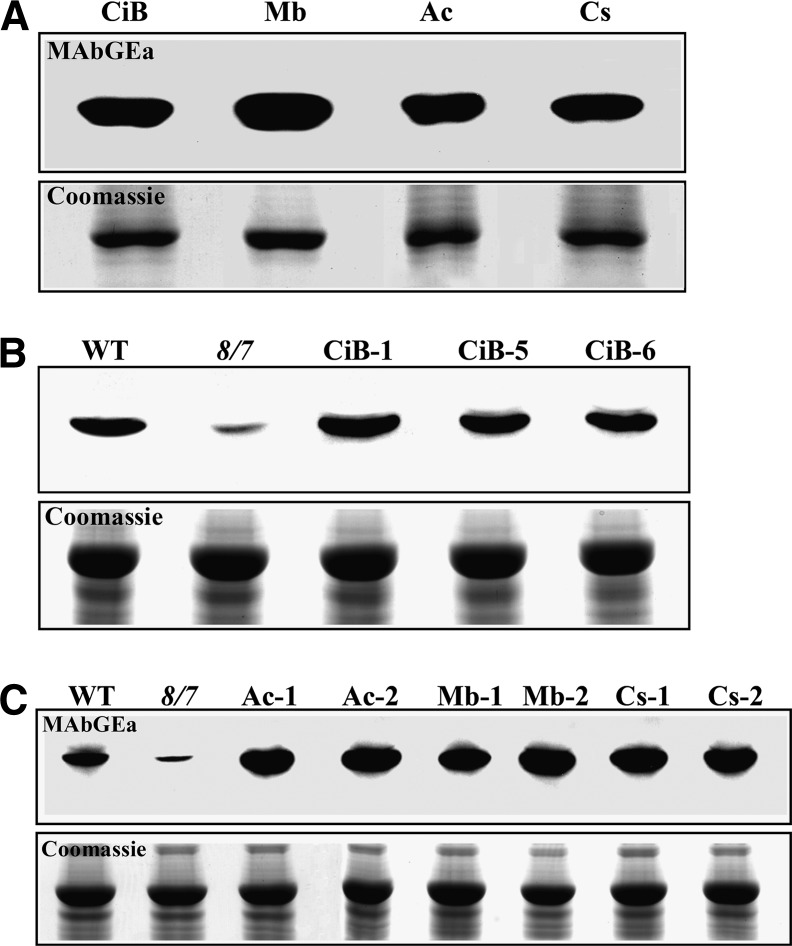
Figure 9.
Immunoblot Analysis of C. intestinalis and Protist Actin Expression in act8-2 act7-4 Plants.
(A) Reactivity of MAbGEa with C. intestinalis cytoplasmic-like actin ACTB1 (CiB) and diverse protist actins. Ac, A. castellanii; Mb, M. brevicollis; Cs, C. scutata. About 1 μg of crude recombinant actin was loaded per lane.
(B) Expression of Ci-ACTB1 in act8-2 act7-4 plants. Three independent lines are shown. WT, the wild type.
(C) Expression of protist actin proteins in act8-2 act7-4 plants. Two independent lines are shown for each protist actin examined.
DISCUSSION
Mounting evidence from previous studies supported the idea that the divergent actin protein variants encoded in multicellular eukaryotes evolved distinct roles essential to cell and organismal development. Contrary to this view, we have shown here that several deuterostome cytoplasmic actin variants and protist actins are functionally indistinguishable from plant vegetative actins in their ability to suppress multiple developmental defects in Arabidopsis mutants lacking vegetative actins. Earlier genetic suppression studies generally resulted in lethality or weak suppression, perhaps because they relied upon the rescue of defects in more specialized actins, such as mouse and Drosophila muscle actins or the relatively divergent budding yeast actin. By contrast, this study examined combinations of animal cytoplasmic actin, plant vegetative actin, and protist actin sequences that are similarly involved in the control of spatial cell development. We can account for the successful suppression results observed in this study only if the suppressing actins are essentially indistinguishable in their cellular functions from the vegetative plant actins. Our data lend support to a new hypothesis that during their independent evolution, cytoplasmic deuterostome actin variants and vegetative plant actin variants inherited their functional competence for spatial development from an ancestral protist actin.
Although Arabidopsis encodes eight actin variants, only the three vegetative actins are predominantly expressed throughout the major sporophytic phase of plant development. We demonstrated earlier by ectopic expression and suppression studies that these vegetative actins are functionally distinct from the reproductive class of actins expressed mainly in pollen and ovules (Kandasamy et al., 2002, 2007). Moreover, genetic suppression studies suggest that At-ACT2 and At-ACT8 can substitute for each other and for At-ACT7 function, but At-ACT7 can only partially support At-ACT2 and At-ACT8 functions in the regulation of root hair tip growth, thus providing evidence for both functional redundancy and divergence, even among the plant vegetative actin variants (Kandasamy et al., 2009). Here, we have shown that the three plant vegetative actins are functionally equivalent to at least three animal cytoplasmic actins and three protist actins, as these nonplant actins are able to fully rescue all the organ development phenotypes of plant actin double mutants. However, unlike the protist actins that support very efficiently the highly specialized tip growth of root hairs, the human and C. intestinalis cytoplasmic actins are only able to partially rescue root hair growth. Thus, the cytoplasmic actins behaved very similarly to vegetative At-ACT7 in supporting only moderate growth of root hairs, although all of them completely rescued plant development. Perhaps At-ACT7 and the animal cytoplasmic actins have partially lost their ability to support rapid cell tip growth and elongation of specialized cells like root hairs, while the protist actins have preserved this property.
It is surprising that Hs-ACTB and Hs-ACTG1, which differ in four N-terminal residues and show dramatic ion-dependent (e.g., Ca2+ and Mg2+) polymerization differences (Bergeron et al., 2010) and apparently serve different functions in humans, are both capable of suppressing all the plant developmental phenotypes. The only distinction between these two human cytoplasmic actins is minor differences in the suppression of root hair cell growth, which is known to be Ca2+ dependent (Wymer et al., 1997; Fan et al., 2011). Hs-ACTB induces root hair growth 10 to 20% better than Hs-ACTG1, consistent with HsACTB being more Ca2+ responsive than Hs-ACTG1. This differential suppression of root hair growth among these two cytoplasmic actins suggests that unlike most of the plant organs or tissues, these specialized root cells are able to recognize the functional differences in these two human sequences. This difference also may be due to actin binding protein interaction with the N terminus or allosteric effects of the N terminus on the rest of the molecule.
The functional homology observed among animal cytoplasmic, plant vegetative, and extant protist actins, as revealed by our positive developmental suppression data, fits well with what has been described as nonprogressive sequence evolution, where ancient protein structures and functions are conserved but the protein sequences are not (Castrodeza, 1979). As plant vegetative actins evolved from common ancestral sequences, they do not appear to have evolved many new functions that are not present in some protist actins or deuterostome cytoplasmic actins. Several observations support this model. First, phylogenetic clustering of the actin sequences examined demonstrated that the animal cytoplasmic actins and two of the protist actin sequences (i.e., Mb-ACT and Ac-ACT1), which suppressed loss of plant actin function, are quite distinct in their sequences from the plant actins (Figure 1B; see Supplemental Figure 2 online). Second, in both animal and yeast actins, a functional role has been shown for the N-terminal region containing several acidic amino acids (Lasa et al., 1997; Hinz et al., 2002; Wong et al., 2002; Lu et al., 2005). Therefore, we were surprised to observe that the animal cytoplasmic actins (i.e., Hs-ACTG1, Hs-ACTB, and Ci-ACTB1) and the protist actins (i.e., Ac-ACT1 and Mb-ACT), which are two amino acids shorter than all plant actins at the N terminus (see Supplemental Figure 2 online), efficiently substituted for plant actins. By contrast, the muscle actins, which are the same length in the N-terminal acidic region as plant actins, did not support plant actin function. In this case, actin function, but not the length at the N terminus, was conserved among actin sequences that rescued mutant phenotypes. Third, changes in protein sequence under a nonprogressive evolutionary model may be viewed as intragenic suppression, where a change in one residue with a deleterious effect is compensated by selection of a second change in sequence that restores the original function. This process of selecting for intragenic suppressor mutations to maintain original structure and function has been proposed to explain paired mutations in HIV-1 protease (Parera et al., 2009), cytochrome c oxidase subunit I (Acín-Pérez et al., 2003), β-subunit of RNA polymerase (Malik et al., 1999), and aminoacyl-tRNA synthetase (Ador et al., 2004) and for the complementary bases in the double-stranded stems of structural RNAs (e.g., tRNA, rRNA, small nuclear RNA, etc.). A comparison of the sequences and three-dimensional structures (see Supplemental Figures 2 and 3 online) of At-ACT2 and Hs-ACTB suggests that their evolution from a common ancestral actin occurred by the codivergence of sets of adjacent amino acids. For example, on one face of actin’s structure depicted in Supplemental Figure 3 online, human β-actin ACTB differs from Arabidopsis ACT2 in five groups of clustered residue changes: Ile-75/Thr-77, Ser-365/Ser-368, Phe-223/Leu-211, Ala-231/Ser-232/Ala-228/Asn-252, and Ser-271/Cys-272/Phe-279. Such clustered changes in sequence account for the majority of their amino acid differences and may represent similar cases of intragenic suppression to maintain wild-type activity. Human skeletal muscle actin ACTA1, which did not suppress plant actin functions, is similarly divergent in many pairs or small sets of adjacent residues, perhaps adjusting for distinct new muscle functions, while maintaining other actin functions, such as nucleotide binding and exchange. Previous studies examining the ability of Drosophila cytoplasmic actin 5C to suppress the loss of flight muscle actin 88F indicated that the combined activities of a number of sequence changes comprised the functional differences of these two variants (Fyrberg et al., 1998). Finally, in spite of the sequence divergence of the cytoplasmic animal and protist actins from the vegetative plant actins, they must have conserved appropriate strength and specificity to their binding of many classes of actin binding proteins. However, the plant actin binding proteins are all quite divergent from those in other kingdoms. For example, plant profilins and actin depolymerizing factors (cofilins) are only 15 and 31% identical in protein sequence to their closest human homologs. Yet the efficient function of animal cytoplasmic actins in plants demonstrates that they must interact appropriately with plant profilins and actin depolymerizing factors.
Although the muscle actins differed by 10 to 14% (37 to 52 amino acid residues) from the plant sequences and the animal cytoplasmic and protist actins differed by only 7 to 13% (Table 1), the suppressing sequences are not phylogenetically much closer to the plant sequences (Figure 1B). However, among the suppressing actins, we found a few conserved amino acid residues that distinguished them from the nonsuppressing actins (see Supplemental Figure 2 online). These residues could be essential to actin’s conservation of competence for spatial development. Based on the residue numbering for At-ACT2, Met-18, Val-78, Thr-164, Leu-178, Thr-203, and Val-288 were perfectly conserved among the nonplant actins that suppressed the act8-2 act7-4 developmental phenotypes or partially suppressed root hair elongation in act2-1 act8-2, while these residues had diverged in the muscle actins. One of these residues, Val-78, is an Ile (Ile-78) in the deuterostome muscle actins we examined and in the protistome Drosophila flight muscle actin 88F. Conversion of Ile-78 in flight muscle actin to Val-78 (i.e., the cytoplasmic animal actin residue) resulted in a flightless phenotype showing the importance of this residue to muscle actin function (Fyrberg et al., 1998). However, the role of Val-78 in supporting the developmental activities of nonmuscle actins is not known. We will test a number of mutant actins in the plant system to determine the importance of these and other residues for cell growth and organ developmental functions.
Two human muscle actins failed to support root organ development and growth in the act8-2 act7-4 mutant, although there was a minor improvement in shoot morphology. Additionally, the muscle actins poorly supported root hair cell tip growth in the act2-1 act8-2 mutant. Thus, there can be little doubt that muscle actins in the deuterostome lineage lost their competence for spatial development. While we can only speculate as to the reasons for this lack of conserved function, a few aspects of their divergence seem particularly worthy of mention. The deuterostome muscle actins are highly specialized in their cellular functions (Khaitlina, 2001) and more divergent in sequence than cytoplasmic actin, as revealed by the longer branch lengths for muscle actins in some phylogenetic analyses of actin sequences (Figure 1B). Cytoplasmic actins are expressed in all cells, including muscle cells, while the muscle actins are all tissue specific. Knockout mice lacking cytoplasmic γ-actin ACTG1 initially develop what appear to be normal skeletal muscles but acquire a progressive myopathy with the subsequent degeneration of skeletal muscle fibers (Sonnemann et al., 2006). Thus, while many cells and tissues develop normally using only cytoplasmic actins, muscle tissues require both cytoplasmic and muscle actin variants, suggesting muscle actins may no longer be competent in fulfilling all cellular and developmental functions. Moreover, the folding of newly synthesized actin into a functional monomer is a chaperone-mediated process. The inability of muscle actins to suppress plant actin function could be due to the divergence of plant chaperone machinery and inappropriate folding into active molecules. For example, a yeast in vitro actin folding system will assemble functional human cytoplasmic ACTB but could not fold functional muscle-specific ACTA1 (Altschuler et al., 2009). Changing Hs-ACTA1 Asn-299 to the Val residue found in yeast ACT1 allowed normal folding of Hs-ACTA1 in this system. Asn-299 is a deuterostome muscle-specific actin residue, while cytoplasmic animal and two protist actins we examined have Thr, and plants and the green alga protist have an Ile in this position. Although the muscle actins were well expressed in plants and equally soluble with the cytoplasmic actins in the low salt extraction buffers used for immunoblot analysis in this study, it will be important to confirm the normal folding of muscle actins expressed in plant cells. Moreover, we are in the process of examining the organization of the actin cytoskeleton in the leaf and root tissues of muscle actin expressing plants that are transformed with the fluorescent fimbrin reporter ABD2-GFP to see whether and how these actins are integrated into the actin filaments in these mildly suppressed and nonsuppressed cell types.
Human cytoplasmic actins appear to function normally in the Arabidopsis model system. It would be interesting to see whether any of the vegetative plant actins or the ancestral protist actins can function in animals and suppress the embryo lethal phenotype of mice lacking ACTB (Shawlot et al., 1998; Bunnell et al., 2011) or the delayed embryonic development and poor survival of mice lacking ACTG1 (Bunnell and Ervasti, 2010). Moreover, because the human cytoplasmic actins are functional in Arabidopsis, missense mutations in these human actins that are associated with a large number of diseases, including blindness, deafness, myopathies, immune deficiency, and aberrant kidney function, could be easily tested in this plant model. For example, the influence of dominant missense mutations in β-actin on the structure of lamellapodia in lymphoblasts (Procaccio et al., 2006) and in γ-actin on the maintenance of stereocilia in the ear (Morín et al., 2009) might be studied in Arabidopsis root hairs. By contrast, examining actin functions using transgenic mouse models would be orders of magnitude more expensive and time consuming. Our positive suppression data indicate that the human cytoplasmic actins and some protist actins have the necessary interactions with a variety of plant actin binding proteins to conduct normal spatial cellular development. However, a significant unresolved issue raised by our results is how the diverse actin functions necessary to support multicellular development could have evolved in and are still maintained in single-celled protists.
METHODS
Plant Material and Generation of Transgenic Plants
Arabidopsis thaliana wild-type (Wassilewskija or Columbia), act2-1 act8-2 and act8-2 act7-4 double null mutants (Kandasamy et al., 2009), and various suppression lines were grown at 22°C with 16 h of light and 8 h darkness. Root morphologies and root hair cell growth were examined on sterile plants grown vertically on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) solidified with 1% agar, and aboveground plant morphologies were observed on soil-grown plants. Fourteen-day-old plants were used for examination of lateral root formation and root architecture, and 24-d-old plants were used for quantitative analysis of leaf size. Roots and root hairs from 25 and 10 T2 generation seedlings (3 d old) of each plant line were examined to quantify root length and root hair length, respectively.
To induce adventitious roots, leaves from sterile seedlings were cultured on solid MS medium supplemented with 0.5 mg/L naphthalene acetic acid and 0.33mg/L indole-3-acetic acid for 4 d. Leaf explants were then transferred to MS plates without hormones and incubated vertically for 6 d for adventitious root growth measurements. For suppression studies, the double mutant plants were transformed with various constructs by the Agrobacterium tumefaciens–mediated floral dip method (Clough, 2005). Fifteen to 20 independent transformed lines in each mutant genetic background were isolated for every transgene by plating sterilized seeds on medium supplemented with suitable antibiotics.
Comparison of Actin Variant Sequences
The actin protein sequences examined include At-ACT2, At-ACT8, At-ACT1, and At-ACT7 (Arabidopsis); Hs-ACTB, Hs-ACTG1, Hs-ACTA1, and Hs-ACTA2 (Homo sapiens); Ci-ACTB1 (Ciona intestinalis); Mb-ACT (Monosiga brevicollis); Ac-ACT1 (Acanthamoeba castellanii); and Cs-ACT (Coleochaete scutata). The previously uncharacterized C. intestinalis sequence Ci-ACTB1 was selected from the C. intestinalis family of actin sequences based on its similarity to human ACTB. The unrooted neighbor-joining tree presented in Figure 1B was constructed using MEGA4 (Tamura et al., 2007). The alignment of the various actin protein sequences was performed using PileUp in GCG version 11.1 (Accelrys), and the multiple-alignment output was made using BOXSHADE version 3.31 C.
Plasmid Construction
The native cDNAs encoding the actin proteins described above (Hs-ACTB, Hs-ACTG1, Hs-ACTA1, Hs-ACTA2, Ci-ACTB1, Mb-ACT, Ac-ACT1, and Cs-ACT) were synthesized and cloned into pUC57 by GenScript. Each of these constructs was then moved into the NcoI/BamHI sites of the Arabidopsis ACTIN2 vector (ACT2pt, A2p) for ubiquitous expression in mutant plants to study suppression. This fragment was also moved into pET15b from EMD4Biosciences for actin protein expression in bacteria to examine the reactivity of the antiactin monoclonal antibodies with different actin variants. The various steps involved in cloning the constructs into ACT2pt were identical to those described previously for the A2P:A2 construct (Kandasamy et al., 2002). The expression plasmids were mobilized into the Agrobacterium strain C58C1 before plant transformation. The actin clones in pET15b were transformed into the expression host BL21 Star (DE3) from Invitrogen for recombinant actin protein expression as described earlier (Kandasamy et al., 1999).
Immunoblot Analysis
To detect actin on immunoblots, total protein from 50 mg of frozen leaf samples was ground frozen in liquid nitrogen and extracted in 250 μL of extraction buffer containing 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM EDTA, and protease inhibitor cocktail (Roche Diagnostics; one tablet/10 mL). After centrifugation, the supernatant was mixed 1:1 with 2× sample buffer (O’Farrell, 1975) and boiled (5 min), and ∼20 μL was loaded per well (i.e., ∼25 μg protein) on a 10% SDS-polyacrylamide gel. The human, C. intestinalis, and protist actins on immunoblots were detected with a mouse monoclonal antibody MAbGEa (Thermo Scientific; MA1-744), which recognized all the eukaryotic actins equally (Kandasamy et al., 1999). Moreover, an anti-HsACT2 antibody (MAbHsACTA2) produced in mouse (Sigma-Aldrich; 4A8-2H3) was used to confirm the expression levels of human muscle actins. Both antibodies were used at a dilution of 1:500 in the blocking solution containing TBST (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20), 10% goat serum, and 5% dry milk. Purified rabbit skeletal muscle actin (Hs-ACTA1) and human cytoplasmic β-actin (Hs-ACTB) that were used for control experiments to show equal reactivity of MAbGEa with both these cytoplasmic, and muscle actins were obtained from Cytoskeleton, Inc. The crude bacterial extracts containing recombinant human (Hs-ACTG1 and Hs-ACTA2), C. intestinalis (Ci-ACTB1), and protist (Ac-ACT1, Mb-ACT, and Ce-ACT) actins made in Escherichia coli were assayed on immunoblots to confirm the reactivity of the antiactin antibodies. Coomassie Brilliant Blue staining of duplicate gels was used to monitor equal loading of proteins and adjust loading if necessary.
Microscopy
Roots used for the examination of the cellular organization and cell lineages at the apex were fixed with 4% paraformaldehyde in 50 mM PIPES buffer, pH 7.0, containing 5 mM EDTA, 1 mM MgCl2, 0.5% casein, and 0.05% Triton X-100 for 30 min to 1 h. The root segments were stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.1 μg/mL) before observation with a Leica fluorescence microscope (Kandasamy et al., 2009). For root and root hair length measurements, 3-d-old seedlings were photographed using a Leica stereomicroscope fitted with a color digital camera.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NP_188508.1, NP_175350, NP_001031504.1, NP_196543.1 (Arabidopsis), P60709.10, CAA27723.1, NP_001091.1, NP_001604.1 (H. sapiens), NP_001027674.1 (C. intestinalis), XP_001747496.1 (M. brevicollis), P02578.1 (A. castellanii), and 065315.1 (C. scutata).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Suppression of Root and Root Hair Growth Phenotypes of Arabidopsis Double Mutants by Human and C. intestinalis Animal and Protist Actins.
Supplemental Figure 2. Comparison of the Sequence of Diverse Actin Variants.
Supplemental Figure 3. Comparison of the Structure of Arabidopsis and Human Actin Variants.
Supplemental Table 1. Fraction of Amino Acid Differences among the Actins Examined in This Study.
Supplemental Data Set 1. Text File of the Actin Sequences Used for Construction of the Phylogenetic Tree Shown in Figure 1B.
Supplementary Material
Acknowledgments
We thank Mark Farmer, Jessica Kissinger, and Jenna Oberstaller for their assistance in the consideration of protists and Nancy Manley for her helpful comments concerning evolution and development throughout this project. We acknowledge the University of Georgia’s Advanced Computing Resource Center for their facilities and technical expertise. This study was supported by a grant from the National Institutes of Health (GM36397-25) to R.B.M. and by National Institutes of Health Genetics Training Grant GM 07103-35 to E.R.
AUTHOR CONTRIBUTIONS
M.K.K. and R.B.M. designed the experiments and wrote the article. M.K.K. and E.C.M. performed the laboratory research and analyzed the data. E.R. contributed to the construction of actin phylogenetic trees.
Glossary
- MS
Murashige and Skoog
- DAPI
4′,6-diamidino-2-phenylindole
References
- Acín-Pérez R., Bayona-Bafaluy M.P., Bueno M., Machicado C., Fernández-Silva P., Pérez-Martos A., Montoya J., López-Pérez M.J., Sancho J., Enríquez J.A. (2003). An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum. Mol. Genet. 12: 329–339 [DOI] [PubMed] [Google Scholar]
- Ador L., Jaeger S., Geslain R., Martin F., Cavarelli J., Eriani G. (2004). Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase. Biochemistry 43: 7028–7037 [DOI] [PubMed] [Google Scholar]
- Altschuler G.M., Dekker C., McCormack E.A., Morris E.P., Klug D.R., Willison K.R. (2009). A single amino acid residue is responsible for species-specific incompatibility between CCT and alpha-actin. FEBS Lett. 583: 782–786 [DOI] [PubMed] [Google Scholar]
- Anderson I.J., Watkins R.F., Samuelson J., Spencer D.F., Majoros W.H., Gray M.W., Loftus B.J. (2005). Gene discovery in the Acanthamoeba castellanii genome. Protist 156: 203–214 [DOI] [PubMed] [Google Scholar]
- Asmussen M.A., Gilliland L.U., Meagher R.B. (1998). Detection of deleterious genotypes in multigenerational studies. II. Theoretical and experimental dynamics with selfing and selection. Genetics 149: 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron S.E., Zhu M., Thiem S.M., Friderici K.H., Rubenstein P.A. (2010). Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J. Biol. Chem. 285: 16087–16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V., Reedy M.C., Sauder U., Kammerer R.A., Aebi U., Schoenenberger C. (1999). Substitution of flight muscle-specific actin by human (beta)-cytoplasmic actin in the indirect flight muscle of Drosophila. J. Cell Sci. 112: 3627–3639 [DOI] [PubMed] [Google Scholar]
- Bryan K.E., Wen K.K., Zhu M., Rendtorff N.D., Feldkamp M., Tranebjaerg L., Friderici K.H., Rubenstein P.A. (2006). Effects of human deafness gamma-actin mutations (DFNA20/26) on actin function. J. Biol. Chem. 281: 20129–20139 [DOI] [PubMed] [Google Scholar]
- Bunnell T.M., Burbach B.J., Shimizu Y., Ervasti J.M. (2011). β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell 22: 4047–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell T.M., Ervasti J.M. (2010). Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton (Hoboken) 67: 564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrodeza C. (1979). Non-progressive evolution, the Red Queen hypothesis, and the balance of nature. Acta Biotheor. 28: 11–18 [DOI] [PubMed] [Google Scholar]
- Clough S.J. (2005). Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286: 91–102 [DOI] [PubMed] [Google Scholar]
- Crawford K., Flick R., Close L., Shelly D., Paul R., Bove K., Kumar A., Lessard J. (2002). Mice lacking skeletal muscle actin show reduced muscle strength and growth deficits and die during the neonatal period. Mol. Cell. Biol. 22: 5887–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 103: 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries M.E., Kelvin A.A., Xu L., Ran L., Robinson J., Kelvin D.J. (2006). Defining the origins and evolution of the chemokine/chemokine receptor system. J. Immunol. 176: 401–415 [DOI] [PubMed] [Google Scholar]
- Donnell A.M., Doi T., Hollwarth M., Kalicinski P., Czauderna P., Puri P. (2008). Deficient alpha-smooth muscle actin as a cause of functional intestinal obstruction in childhood. Pediatr. Surg. Int. 24: 1191–1195 [DOI] [PubMed] [Google Scholar]
- Fan J.L., Wei X.Z., Wan L.C., Zhang L.Y., Zhao X.Q., Liu W.Z., Hao H.Q., Zhang H.Y. (2011). Disarrangement of actin filaments and Ca2+ gradient by CdCl2 alters cell wall construction in Arabidopsis thaliana root hairs by inhibiting vesicular trafficking. J. Plant Physiol. 168: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin L.K., et al. (2010). The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140: 631–642 [DOI] [PubMed] [Google Scholar]
- Fyrberg E.A., Fyrberg C.C., Biggs J.R., Saville D., Beall C.J., Ketchum A. (1998). Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 36: 271–287 [DOI] [PubMed] [Google Scholar]
- Gilliland L.U., Kandasamy M.K., Pawloski L.C., Meagher R.B. (2002). Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol. 130: 2199–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland L.U., McKinney E.C., Asmussen M.A., Meagher R.B. (1998). Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual Arabidopsis actin genes. Genetics 149: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland L.U., Pawloski L.C., Kandasamy M.K., Meagher R.B. (2003). Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 33: 319–328 [DOI] [PubMed] [Google Scholar]
- Hedges S.B., Blair J.E., Venturi M.L., Shoe J.L. (2004). A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 4: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower R.C., Meagher R.B. (1986). The molecular evolution of actin. Genetics 114: 315–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B., Gabbiani G., Chaponnier C. (2002). The NH2-terminal peptide of alpha-smooth muscle actin inhibits force generation by the myofibroblast in vitro and in vivo. J. Cell Biol. 157: 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., Burgos-Rivera B., McKinney E.C., Ruzicka D.R., Meagher R.B. (2007). Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19: 3111–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., Gilliland L.U., McKinney E.C., Meagher R.B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (1999). The late pollen-specific actins in angiosperms. Plant J. 18: 681–691 [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (2002). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (2009). A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21: 701–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson R., Aspenström P., Byström A.S. (1991). A chicken beta-actin gene can complement a disruption of the Saccharomyces cerevisiae ACT1 gene. Mol. Cell. Biol. 11: 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.J. (2007). Genomics. Deep questions in the tree of life. Science 317: 1875–1876 [DOI] [PubMed] [Google Scholar]
- Khaitlina S.Y. (2001). Functional specificity of actin isoforms. Int. Rev. Cytol. 202: 35–98 [DOI] [PubMed] [Google Scholar]
- King N., et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451: 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., et al. (1997). Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc. Natl. Acad. Sci. USA 94: 4406–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I., Gouin E., Goethals M., Vancompernolle K., David V., Vandekerckhove J., Cossart P. (1997). Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. EMBO J. 16: 1531–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.Y., Ferrenberg A.M., Gilliland L.U., Meagher R.B., Asmussen M.A. (2003). Detection of deleterious genotypes in multigenerational studies. III. Estimation of selection components in highly selfing populations. Genet. Res. 82: 41–53 [DOI] [PubMed] [Google Scholar]
- Lu X., Bryant M.K., Bryan K.E., Rubenstein P.A., Kawai M. (2005). Role of the N-terminal negative charges of actin in force generation and cross-bridge kinetics in reconstituted bovine cardiac muscle fibres. J. Physiol. 564: 65–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik T., Ahmad K., Buyukuslu N., Cromie K., Glass R.E. (1999). Intragenic suppression of trans-dominant lethal substitutions in the conserved GEME motif of the beta subunit of RNA polymerase: Evidence for functional cooperativity within the C-terminus. Genes Cells 4: 501–515 [DOI] [PubMed] [Google Scholar]
- Meagher R.B. (1995). The impact of historical contingency on gene phylogeny: Plant actin diversity. In Evolutionary Biology, M. Hecht, R. MacIntyre, and M. Clegg, eds (New York: Plenum Press), pp. 195–215 [Google Scholar]
- Meagher R.B, Kandasamy M.K, King L. (2011). Actin functions in the cytoplasmic and nuclear compartments. In Plant Cytoskeleton. Advances in Plant Biology, Vol. 2, B. Liu, ed (Dordrecht, The Netherlands: Springer Science/Business Media), pp. 3–32 [Google Scholar]
- Meagher R.B., McKinney E.C., Vitale A.V. (1999). The evolution of new structures: Clues from plant cytoskeletal genes. Trends Genet. 15: 278–284 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morín M., Bryan K.E., Mayo-Merino F., Goodyear R., Mencía A., Modamio-Høybjør S., del Castillo I., Cabalka J.M., Richardson G., Moreno F., Rubenstein P.A., Moreno-Pelayo M.A. (2009). In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum. Mol. Genet. 18: 3075–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Plant Physiol. 15: 473–497 [Google Scholar]
- Nunoi H., Yamazaki T., Tsuchiya H., Kato S., Malech H.L., Matsuda I., Kanegasaki S. (1999). A heterozygous mutation of beta-actin associated with neutrophil dysfunction and recurrent infection. Proc. Natl. Acad. Sci. USA 96: 8693–8698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H. (1975). High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250: 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- Parera M., Perez-Alvarez N., Clotet B., Martínez M.A. (2009). Epistasis among deleterious mutations in the HIV-1 protease. J. Mol. Biol. 392: 243–250 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte P., Gunning P., Blau H., Kedes L. (1983). Human actin genes are single copy for alpha-skeletal and alpha-cardiac actin but multicopy for beta- and gamma-cytoskeletal genes: 3′ untranslated regions are isotype specific but are conserved in evolution. Mol. Cell. Biol. 3: 1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccio V., et al. (2006). A mutation of beta -actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am. J. Hum. Genet. 78: 947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildmeyer L.A., Braun R., Taffet G., Debiasi M., Burns A.E., Bradley A., Schwartz R.J. (2000). Impaired vascular contractility and blood pressure homeostasis in the smooth muscle alpha-actin null mouse. FASEB J. 14: 2213–2220 [DOI] [PubMed] [Google Scholar]
- Shawlot W., Deng J.M., Fohn L.E., Behringer R.R. (1998). Restricted beta-galactosidase expression of a hygromycin-lacZ gene targeted to the beta-actin locus and embryonic lethality of beta-actin mutant mice. Transgenic Res. 7: 95–103 [DOI] [PubMed] [Google Scholar]
- Sonnemann K.J., Fitzsimons D.P., Patel J.R., Liu Y., Schneider M.F., Moss R.L., Ervasti J.M. (2006). Cytoplasmic gamma-actin is not required for skeletal muscle development but its absence leads to a progressive myopathy. Dev. Cell 11: 387–397 [DOI] [PubMed] [Google Scholar]
- Sparrow J.C., Nowak K.J., Durling H.J., Beggs A.H., Wallgren-Pettersson C., Romero N., Nonaka I., Laing N.G. (2003). Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul. Disord. 13: 519–531 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Blanchoin L. (2006). Actin dynamics: Old friends with new stories. Curr. Opin. Plant Biol. 9: 554–562 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Wong W.W., Gerson J.H., Rubenstein P.A., Reisler E. (2002). Thin filament regulation and ionic interactions between the N-terminal region in actin and troponin. Biophys. J. 83: 2726–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer C.L., Bibikova T.N., Gilroy S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12: 427–439 [DOI] [PubMed] [Google Scholar]
- Zhu M., Yang T., Wei S., DeWan A.T., Morell R.J., Elfenbein J.L., Fisher R.A., Leal S.M., Smith R.J., Friderici K.H. (2003). Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26). Am. J. Hum. Genet. 73: 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A., Lang D., Richardt S., Frank W., Reski R., Rensing S.A. (2007). Dating the early evolution of plants: Detection and molecular clock analyses of orthologs. Mol. Genet. Genomics 278: 393–402 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.