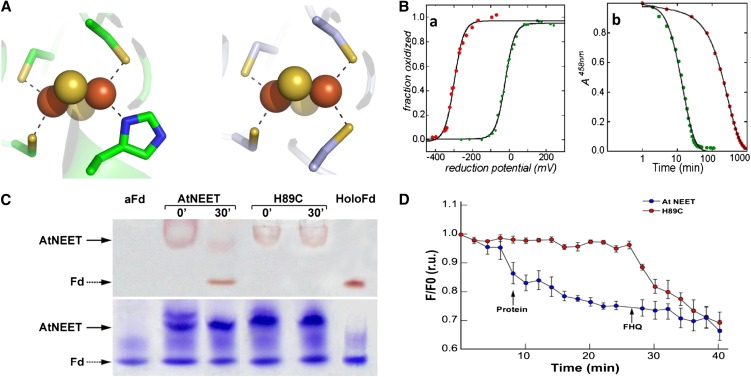
Figure 11.
Characterization of the 2Fe-2S Cluster in the At-NEET H89C Mutant Protein.
(A) Comparison of the cluster binding structures of wild-type At-NEET (pdb ID code 3S2Q; left) and the H89C mutant (pdb ID code 3S2R; right). As the overall secondary structure of the mutant remains the same as that of the wild type (see Supplemental Figure 5 online), the major structural changes are observed in the immediate vicinity of the mutation.
(B) Biophysical characteristics of the At-NEETH89C 2Fe-2S cluster. (a) Decrease in redox potential. Measurement of the fraction oxidized versus reduction potential showing that the redox potentials (given by reduction potential at which the 2Fe-2S clusters are half reduced) differ by ∼300 mV in magnitude with the H89C mutant being lower (red trace versus green trace of the wild-type At-NEET). (b) Increase in cluster stability. A time course of cluster stability at pH 6.0 showing that the 2Fe-2S cluster of the H89C mutant is more stable than the wild type by approximately an order of magnitude (red trace versus green trace of the wild-type At-NEET).
(C) Cluster transfer analysis of wild-type At-NEET and the H89C mutant. At-NEET or the At-NEETH89C mutant were incubated with aFd for 30 min in a transfer assay buffer and analyzed by native-PAGE. Gels are shown before (top) and after (bottom) staining with Coomassie blue. The red-colored bands of the Fe-S cluster–containing proteins are indicated by arrows.
(D) Transfer of labile Fe/FeS from wild-type or mutated At-NEET to RPA-labeled permeabilized HEK293 cells. RPA fluorescence was measured 20 min after the addition of 10 µM wild-type (blue) or At-NEETH89C (red) proteins. Data are represented as mean ± se (n = 10). r.u., relative units.