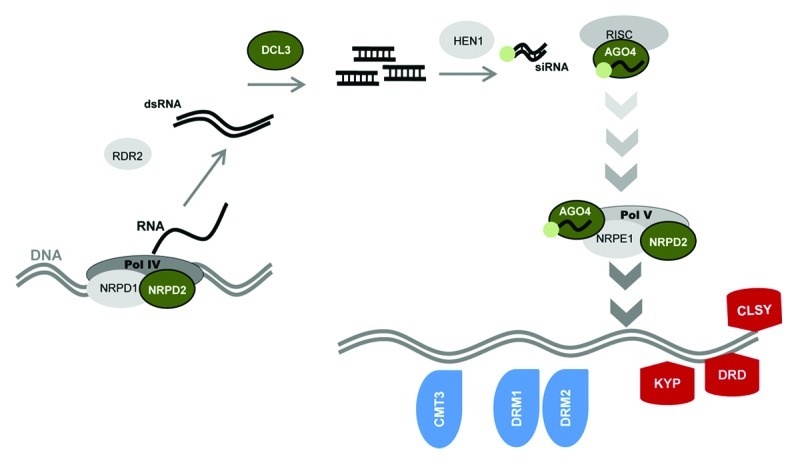
Figure 1. Model of the RNA-directed DNA methylation (RdDM) pathway, adapted from.25,26 Multi-subunit DNA-dependent RNA Polymerase IV (Pol IV) generates single stranded RNAs, which are used as a template by RNA-dependent RNA polymerase 2 (RDR2) to generate double-stranded RNAs (dsRNAs). DICER-LIKE 3 (DCL3) processes dsRNAs into 24 nucleotide RNAs that become methylated by the RNA-methyltransferase HEN1 and subsequently recruited by the AGO4-RISC complex to targeted genomic sites via interaction with the DNA-dependent RNA Polymerase V (Pol V). Pol V is required for DNA methyltransferase activity by DRM1, DRM2 and CMT3 and chromatin remodelling enzymes DRD, CLSY and KYP. The RdDM pathway targets cytosine (C) methylation at non-CpG sites. Signaling components affected in the mutants tested for transgenerational SAR are shown in color; green: enzymes in the siRNA machinery; red: chromatin remodelling enzymes; blue: DNA methyltransferase enzymes.
