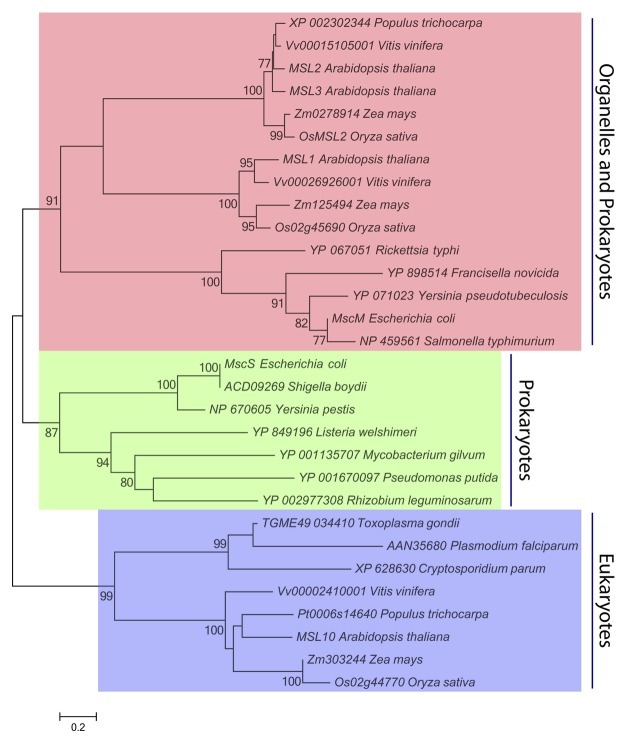
Figure 1. MscS Family Members in Organelles and Intracellular Pathogens. Thirty MscS homologs from 14 species of bacteria and 5 species of plants were identified by Aramemnon cluster analysis (http://aramemnon.botanik.uni-koeln.de/index.ep), previously published analyses,19,25 or inclusion in the Ortho MCL database (http://orthomcl.org/cgi-bin/OrthoMclWeb.cgi). The MscS-like region of each protein sequence was determined by InterProScan,26 and aligned using ClustalX27 with a multiple sequence alignment gap opening penalty of 3.0 and an extension penalty of 1.8. A total of 326 positions, including gaps, were present in the final data set. Evolutionary relationships were inferred by Neighbor-Joining28 and JTT distance matrix methods29 using MEGA5 software. The percentage of trees in which the associated taxa clustered together in the bootstrap test (n = 2000 replicates) are shown next to the branches.30 Only significant bootstrap values (greater than 75%) are shown, and branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. Scale bar, 0.2 amino acid substitutions per site.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.