SUMMARY
Iron can regulate biofilm formation via non-coding small RNA (sRNA). To determine if iron-regulated sRNAs are involved in biofilm formation by the periodontopathogen Aggregatibacter actinomycetemcomitans, total RNA was isolated from bacteria cultured with iron supplementation or chelation. Transcriptional analysis demonstrated that the expression of four sRNA molecules (JA01-JA04) identified by bioinformatics was significantly up-regulated in iron-stressed medium compared to iron-rich medium. A DNA fragment encoding each sRNA promoter was able to titrate E. coli Fur from a Fur-repressible reporter fusion in an iron uptake regulator titration assay. Cell lysates containing recombinant AaFur shifted the mobility of sRNA-specific DNAs in a gel shift assay. Potential targets of these sRNAs, determined in silico, included genes involved in biofilm formation. A. actinomycetemcomitans overexpressing JA03 sRNA maintained a rough phenotype on agar, but no longer adhered to uncoated polystyrene or glass, although biofilm determinant gene expression was only modestly decreased. In summary, these sRNA have the ability to modulate biofilm formation, but their functional targets genes remain to be confirmed.
Keywords: iron, Fur, sRNA, biofilm, regulation, transcription
INTRODUCTION
Aggregatibacter actinomycetemcomitans is a gram-negative, capnophilic bacterium associated with aggressive forms of periodontitis and less commonly with systemic diseases such as endocarditis, atherosclerosis, and brain abscesses (Kaplan et al., 1989, Zambon, 1985). A. actinomycetemcomitans produces flp bundle-forming fimbriae, extrapolymeric substance (EPS), and lipopolysaccharide (LPS), all of which have been implicated in initial colonization and biofilm formation (Inoue et al., 1998, Kachlany et al., 2000, Kaplan et al., 2003, Rosan et al., 1988, Scannapieco et al., 1987, Schreiner et al., 2003, Tomich et al., 2007). Although environmental factors such as nutrient stress, pH, oxygen tension (Haase et al., 2006, Scannapieco et al., 1987) and iron (Fe) concentration (Amarasinghe et al., 2009) have been shown to influence the expression of fimbriae and other biofilm determinants, the molecular mechanisms that regulate biofilm formation are unknown. In other bacterial species, such as Vibrio cholerae and Escherichia coli, biofilm formation is regulated in part by Fe via short non-coding RNA molecules called small regulatory RNA (sRNA) (Mey et al., 2005a, Mey et al., 2005b). We hypothesize that Fe-regulated sRNAs play a role in biofilm formation and dispersion in A. actinomycetemcomitans.
Small regulatory RNAs play an important role in the regulation of bacterial gene expression especially under stress conditions. These sRNA are typically 50–200 nucleotides in length and are not translated into protein (Gottesman, 2004a, Gottesman, 2004b). Most bacterial sRNAs identified so far target mRNAs (in trans) via imperfect sequence complementary base pairing (Gottesman, 2002, Masse et al., 2003). A well-known sRNA molecule in E. coli, RyhB, is repressed by the ferric uptake regulator (Fur) protein in E. coli and is expressed by the cell only during Fe starvation (Gottesman, 2002, Masse et al., 2003). Binding of Fur to a promoter commonly negatively regulates gene transcription. The discovery of RyhB provides a mechanism by which Fur can indirectly increase transcription from certain promoters. Under conditions of Fe limitation, Fur has no binding affinity for the Fur box located near the ryhB promoter, allowing ryhB to be transcribed. RyhB sRNA promotes the degradation of sodB mRNA, a Fe superoxide dismutase, and other target transcripts (Gottesman, 2002, Masse et al., 2003). In Fe-sufficient conditions, Fur assumes a conformation with high affinity for the Fur box in the ryhB promoter. Binding of Fur represses transcription of ryhB, thus preventing degradation of the target mRNAs. This paper reports the presence of several Fe- and Fur-regulated sRNA molecules in A. actinomycetemcomitans that may be involved in biofilm development.
METHODS
Bacterial strains and culture conditions
Strains and plasmids used in this study are listed in Table 1. A. actinomycetemcomitans HK1651 rough phenotype (HK1651R) was cultured for 48–72 h anaerobically (5% CO2, 10 % H2, 85 % N2) at 37°C from frozen stock to plates containing Bacto™ tryptic soy broth (Becton, Dickinson and Co.) supplemented with 0.6% yeast extract, 0.04% sodium bicarbonate (TSBY), and 1.5% Bacto™ agar (Becton, Dickinson and Co.). Broth cultures were grown statically overnight in TSBY at 37°C either in an anaerobic chamber or in a candle extinction jar. The spontaneous nalidixic acid-resistant strain HKR.Nal was obtained by growing a dense mid-log cell suspension on TSBY agar containing 20 μg ml−1 nalidixic acid followed by culture on plates containing 50 μg ml−1 nalidixic acid. TSBY (Fe-sufficient) medium used in routine culturing of A. actinomycetemcomitans contains sufficient Fe for robust growth. Fe-supplemented conditions were achieved by addition of FeCl3 to 300 μM to TSBY just prior to use, while Fe-limited conditions (Fe-chelated) were obtained by addition of the Fe chelator 2, 2′-dipyridyl (Sigma, St. Louis, MI) to 300 μM to the culture medium. Mutant strains of A. actinomycetemcomitans were grown in TSBY supplemented with 40 μg ml−1 kanamycin, and overexpressing strains were grown in TSBY supplemented with 2 μg ml−1 chloramphenicol. E.coli and corresponding plasmids were maintained in Difco LB broth, Lennox (Becton, Dickinson and Co.) supplemented with 50 μg ml−1 kanamycin or 100 μg ml−1 ampicillin, as needed.
Table 1.
List of strains used and engineered in this study.
| Strain | Relevant characteristic(s)1 | Source |
|---|---|---|
| A. actinomycetemcomitans | ||
| HK1651R | Clinical isolate HK1651, genome project; (R) rough colony variant | ATCC2 |
| HKR.Nal | HK1651R with a spontaneous mutation (Nalr) | This work |
| HKR.fur::kan | HKR.Nal with fur::kan mutation; Nalr, Kmr | This work |
| HKR.fur::kan-pJAK16 | HKR.fur::kan with pJAK16; Nalr, Kmr, Cmr | This work |
| HKR.fur::kan-pJAKfur | HKR.fur::kan with pJAK-fur; Nalr, Kmr, Cmr | |
| HKR(pJAK16) | HKR.Nal containing pJAK16; Nalr, Cmr | This work |
| HKR(pJAK-fur) | HKR.Nal containing pJAK-fur; fur overexpression strain; Nalr, Cmr | This work |
| HKR(pJAK16-JA03) | HKR.Nal containing pJAK-JA03; JA03 overexpression strain; Nalr, Cmr | This work |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG phi80lacZΔM15 | Invitrogen |
| BL21(DE3) | fhuA2 (lon) ompT gal (λ DE3) (dcm) ΔhsdS λ DE3 = λ sBamHIo ΔEcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 Δnin5 | Novagen |
| BL21(DE3)(pAaFur) | BL21(DE3) containing pAaFur | This work |
| SK140 | Top10 containing mobility plasmid pRK21761); Kmr | (Kachlany et al., 2000) |
| SK140(pJAK16) | SK140 containing pJAK16; Kmr, Cmr | This work |
| SK140(pJAK-fur) | SK140 containing pJAK16-fur; Kmr, Cmr | This work |
| SK140(pJAK16-JA03) | SK140 containing pJAK16-JA03; Kmr, Cmr | This work |
| H1717 | araD139 Δ(argF-lac) U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR aroB fhuF::λ placMu53, Kmr, Smr, (fhuF-lacZ fusion, with fur) | (Hantke, 1987) |
| H1780 | araD139 Δ(argF-lac) U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR fiu:: λplacMu53, (fiu-lacZ fusion, lacking fur) | (Hantke, 1987) |
| Plasmids | ||
| pGEM-T | TA cloning vector; Ampr | Promega |
| pGEMT-fur | pGEM-T containing HK1651R fur with engineered KpnI, BamHI sites; Ampr | This work |
| pGEMT-fur::kan | pGEM-T containing HK1651R fur::kan with engineered KpnI, BamHI sites; Ampr | This work |
| pGEMT-JA03 | pGEM-T containing HK1651R JA03 with engineered KpnI, BamHI sites; Ampr | This work |
| pGEMT-JA01pro | pGEM-T containing the HK1651R JA01 promoter; Ampr | This work |
| pGEMT-JA02pro | pGEM-T containing the HK1651R JA02 promoter; Ampr | This work |
| pGEMT-JA03pro | pGEM-T containing the HK1651R JA03 promoter; Ampr | This work |
| pGEMT-JA04pro | pGEM-T containing the HK1651R JA04 promoter; Ampr | This work |
| pET-30b | Novagen expression vector; Ampr | Novagen |
| pAaFur | pET-30b with the HK1651R fur gene cloned into the NcoI/NtoI sites; Ampr | This work |
| pRK21761 | RK2 IncP Kmr, Ampr, Tets, Lac+ (tetA::lacZ) oriT1 Mob+ | (Sia et al., 1996) |
| pMB78 | USS-containing cloning vector; suicide vector used for allelic exchange; Ampr | (Bhattacharjee et al., 2007) |
| pMfur | pMB78 containing HK1651R fur; Ampr | This work |
| pMfur.KanEZ | pMfur with insertion in fur gene; Ampr, Kmr | This work |
| pMJA03 | pMB78 containing HK1651R JA03; Ampr | This work |
| pJAK16 | IncQ expression plasmid; Cmr | (Thomson et al., 1999) |
| pJAK-fur | pJAK16 containing HK1651R fur; Cmr | This work |
| pJAK-JA03 | pJAK16 containing HK1651R JA03; Cmr | This work |
| Transposon | ||
| EZ::Tn5<KAN-2> | Tn5 transposon containing a Kmr cassette | Epicentre |
Amp; ampicillin; Cm, chloramphenicol; Km, kanamycin; Tet, tetracycline; r, resistant; s, sensitive; Sm, streptomycin.
ATCC, American Type Culture Collection.
Identification of Fur-regulated sRNA molecules
Several potentially Fur-regulated sRNA molecules were identified in A. actinomycetemcomitans using a bioinformatics approach based on three criteria previously reported to identify sRNA sequences in other bacterial species: (1) sequences present in InterGenic Regions (IGR), (2) sequences ending with a ρ-independent terminator, a stem-loop followed by a run of T nucleotides, and (3) sequences regulated directly by the Fur protein (containing a Fur box in the promoter region) (Wilderman et al., 2004). The A. actinomycetemcomitans HK1651 genome (http://www.genome.ou.edu/act.html) (Roe et al.) was first partitioned into two data sets at the Oral Pathogen Sequence Database (http://www.oralgen.lanl.gov/), one that included intergenic sequences and the other consisting of all the annotated ORFs. Using a simple pattern search based on the degenerate E. coli Fur-box sequence, NAT(A/T)ATNAT(A/T)ATNAT(A/T)ATN (Escolar et al., 1999), several potential Fur-regulated sRNA molecules were identified in the intergenic data set. Next, the RNA structure prediction software Mfold (Version 3.2, http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) (Zuker, 2003) was used to select among the sRNAs for structurally more reliable molecules with enthalpy values ranging from −20 to −38, potential Fur boxes and terminal stem-loop structures immediately followed by a series of T nucleotides (U nucleotide for RNA).
sRNA target prediction
Once potential Fur-regulated sRNA molecules were identified, the software program TargetRNA (Version 1.0, http://cs.wellesley.edu/~btjaden/predictions/aggregatibacter/index.html) (Tjaden et al., 2006) was used to identify potential mRNA targets. The following default parameter settings for TargetRNA were used, as previously described: the individual base-pair model of hybridization scoring was used, the putative terminator stem-loop of sRNAs was removed, the search for base-pair binding in mRNA sequences was restricted to a region from 30 nucleotides upstream of translation initiation to 20 nucleotides downstream of translation initiation, G-U base-pairs were not considered when the hybridization seeds of a minimum number of consecutive base-pairs were determined, and potential base-pair binding interactions were considered significant only if their P value fell below 0.05 (Mellin et al., 2007). Further, mRNA sequences were considered as potential targets only if their predicted interaction with sRNAs contained a hybridization seed of at least eight consecutive base pairs (Mellin et al., 2007).
RNA isolation and analysis
Total RNA was prepared from A. actinomycetemcomitans by a modification of the SDS lysis/CsCl cushion procedure, as previously described (Haase et al., 2003). Briefly, bacterial cells were grown anaerobically to late log to early stationary phase (~21 h), scraped from the vessel walls, and harvested by centrifugation. The cells were washed with cold lysis buffer (50 mM Tris-HCl, pH 7.5), and resuspended in lysis buffer containing RNAprotect™ (Qiagen). The suspension was passed through a 20-gauge needle several times to break up aggregated cells. One milliliter of 20% SDS was added to the cell suspension, then vortexed briefly and incubated at 37°C with vortexing once every minute until the cells were completely lysed. This was followed by addition of 4 g of solid CsCl to the suspension, with slow mixing by gentle inversion for 2 min. An additional 8 ml of cold lysis buffer was added and mixed, and the precipitate removed by centrifugation at 15,000 × g for 10 min. The supernatants were carefully layered onto 4 ml of 5.7 M CsCl cushions. Total RNA was pelleted by ultracentrifugation (Beckman L8-M with a SW28 rotor) at 102,000 × g at 20°C for 28 h. Supernatants were removed by inversion, the pellets dissolved in 100 μl diethyl pyrocarbonate (DEPC)-treated water, 3 volumes 95% ethanol were added and the RNA was pelleted by centrifugation, pellets were reconstituted in 100 μl DEPC-treated water, aliquoted and stored at −70°C. RNA was evaluated for quantity by measurement of absorption at A260 and A280, using a Nanodrop spectrophotometer, and for integrity by use of formaldehyde agarose gel electrophoresis.
RNA isolation for Northern blots
Total RNA was isolated from A. actinomycetemcomitans at ~21 h during late log to early stationary growth phase (OD600 ~0.12). Briefly, cells were grown in 20 ml TSBY in a tissue culture flask (Falcon) for about 24 h at 37°C in a candle jar. Cells were scraped from the vessel walls and pelleted by centrifugation for five min at 6,000 × g at 4°C. Pellets were resuspended in cold lysis buffer (50 mM Tris-HCl, pH 7.5) containing RNAprotect™ (Qiagen), incubated at room temperature for five min prior to centrifugation in swinging bucket rotor at 4,000 × g at 20°C. The RiboPure™-Bacteria Kit (Ambion, Applied Biosystems) was used to recover total RNA according to manufacturer’s instructions with the following modifications. After the addition of lysis buffer (RNAwiz), the pellet was mixed by pipetting up and down several times for one min, and then vortexed for one min to break up microcolonies before adding to a tube containing Zirconia beads. The tubes were placed on a Mini-Beadbeater 8 (BioSpec Products) and pulsed for one min at 4°C followed by resting for one min; the bead beating cycle was repeated a total of three times. Total RNA was recovered from the lysate by extraction with chloroform. To enrich for sRNA, 1/3 volume of 100% ethanol was added to the aqueous layer and mixed thoroughly before applying to a spin column filter from the mirVana™ miRNA Isolation kit (Ambion, Applied Biosystems). Filtrate was applied to another spin column and eluted following manufacturer’s protocol. RNA samples were treated with Turbo-DNase-free (Ambion, Biosystems), quantified on the Nanodrop spectrophotometer, and integrity assessed by agarose gel electrophoresis.
Northern blot
sRNA was isolated from A. actinomycemcomitans as described. sRNA samples (10 μg) were combined with gel loading buffer II (mirVana miRNA kit) and incubated at 95°C for 5 min prior to loading. Gels of 6% AccuGel™ 19:1acrylamide (National Diagnostics) and 7 M urea were prepared in 1X TBE (Tris borate EDTA, National Diagnostics) and prerun in 1X TBE for 1 h at 400 V. After loading, gels were run for ~1 h at 200 V in 1X TBE. RNA size standards ranging from 1,000 to 100 bases (RNA Century-Plus ladder, Applied Biosystems) were used to estimate transcript size. DNA (2 μg) corresponding to each sRNA used to generate the Northern blot probe was used as an additional control. RNA was transblotted (Hoeffer Scientific) to Hybond N+ membrane (Amersham, GE Healthcare) in 1X TBE for 1 h 15 min at 200 V, and UV cross-linked. Blots were prehybridized and hybridized overnight at 42°C in ULTRAHyb® buffer (Ambion, Applied Biosystems) according to the NorthernMax® protocol (Ambion, Applied Biosystems). To generate double-stranded DNA probes complementary to candidate sRNA, chromosomal DNA from strain HK1651R (MasterPure™ DNA Purification Kit, Epicentre) was PCR amplified with primers as listed in (Table 2). Reactions for JA01, JA02, and JA03 were: 2 min at 95°C (1 cycle), followed by 30 cycles of 30 s at 95°C, 1 min at 51°C, 2 min at 72°C, and 10 min at 72°C (1 cycle). An annealing temperature of 56°C was used with JA04 primers. PCR products were column-purified (QIAquick, Qiagen) and quantified using the Nanodrop spectrophotometer. DNA was labeled with biotin using the BrightStar®–Psoralen-Biotin kit (Ambion, Applied Biosystems), and assessed for sensitivity according to manufacturer’s protocol. dsDNA probes were used at 1 pM and denatured prior to use. After hybridization, blots were washed with low stringency and high stringency buffers (NorthernMax®) followed by detection of the biotinylated probes using the BrightStar®–Biodetect™ Kit (Ambion, Applied Biosystems) according to manufacturer’s protocols. Signals were detected by exposure to autoradiography Hyperfilm ECL (Amersham, GE Healthcare).
Table 2.
List of oligonucleotides used in this study.
| Primer Name | Primer sequence (5′-3′)1,2,3,4 |
|---|---|
| Plasmid verification | |
| T7-F (pGEMT vectors) | TAATACGACTCACTATAGGG |
| pTAC-F | TTGACAATTAATCATCGGCTCGTATAAT |
| pJAK16-R | CACACTACCATCGGCGCTAC |
|
| |
| Northern blot probes | |
| JA01-F | ACCGCTAAATAATAGCCAACTGGC |
| JA01-R | CAGTGCACTTGTGCCGATAACCT |
|
| |
| JA02-F | GCACAAGATAATGTGTTGGATGTGA |
| JA02-R | AATGCGATTTTTACCACGCA |
|
| |
| JA03-F | AGCCACGTAATAACTGCTGA |
| JA03-R | TGTTATGCGAAATTAGTGACAAAGT |
|
| |
| JA04-F | ATTCTTGCGAACTTTTTCCGTT |
| JA04-F | TCTCACGAAGAGAGGCAAATAACC |
|
| |
| sRNA & Fur overexpression | |
|
| |
| JA03-KpnI-F | CGGGGTACCCCGTAATGCGAAACGTGCTTGAA; KpnI |
| JA03-BamHI-R | CGCGGATCCGCGCCAACTCCTTTCCTTAATTTGTT; BamHI |
|
| |
| Fur-KpnI-F | CGGGGTACCCCGGCTTAATAGCAAAAAGGAGCTG; KpnI |
| Fur-BamHI-R | CGCGGATCCGCGAGACCGCCTTTTGCATGGT; BamHI |
|
| |
| ActF-194-F | TGACACAGGCAATACGGAAA |
|
| |
| FURTA and EMSA | |
| JA01-pro-F | TTCTACGCAAGGTTGGTTTATTT |
| JA01-pro-R | TGTTATCTACGCACTATATCAAATTCA |
|
| |
| JA02-pro-F | ATTTTGCACAAGATAATGTGTTGG |
| JA02-pro-R | AAATTTGTCCCTTTACACAAATTTTA |
|
| |
| JA03-pro-F | AAAGCCACGTAATAACTGCTGA |
| JA03-pro-R | GAAAACACAGAGTTATTCAAAGGTCA |
|
| |
| JA04-pro-F | TTTTGATTAAAAATTAAGCGTTCG |
| JA04-pro-R | AAGCCTGCGATTTTCCTGTA |
|
| |
| Antibiotic cassette | |
| Km-R | GCAATGTAACATCAGAGATTTTGAG |
|
| |
| TaqMan probes/primers for RT-qPCR | |
| tadV-F | GCGGCCAAGTTTTTTCTTTTTCTT |
| tadV-R | GCAATCCGTTTTCTTTAATTGATTTACG |
| tadV-MGB probe | CCGGATTGGGACTAATT |
|
| |
| pgaC3-F | TGGTTCAAGCCTTAGAGCAAGATC |
| pgaC3-R | CGGTTACGTACACGCGGATTA |
| pgaC3- MGB probe | CCTGTGGTAGCAGCATAT |
|
| |
| rmlB-F | AACGTATTAGAAGATTGGTGCCTGTT |
| rmlB-R | CCTCCCATGACCCAATCGAAA |
| rmlB-MGB probe | AAACCACGCCTTATTC |
|
| |
| JA01-F | TGACCGCTAAATAATAGCCAACTG |
| JA01-R | TGTGCCGATAACCTTAAAATCATTTT |
| JA01-Probe | FAM-CGATTTCTTTGGATGGTTTTCAAGGCACTATAAC-NFQ |
|
| |
| JA03-F | CGTGCTTGAATTTATGCTTACACTTC |
| JA03-R | AACACAGAGTTATTCAAAGGTCACTATTACTAG |
| JA03-Probe | FAM-TGTTACAAGGCTTTATTTCTTCAGTTTTCATCACGTT-NFQ |
|
| |
| JA04-F | TCTTGCGAACTTTTTCCGTTATACT |
| JA04-R | CCTAAGGAACCAATTTTATTAAAACAACTG |
| JA04-Probe | FAM-TGTCATAAATTACGAGCAACT-NFQ |
|
| |
| 16S rRNA-F | ACGCGAAGAACCTTACCTACTCT |
| 16S rRNA-R | CCTAAGGCACAAACCACATCTCT |
| 16S rRNA-MGB probe | CATCCGAAGAAGAACTC |
|
| |
| tadG-F | ATCCCTTCCTTGCCGAGTGT |
| tadG-R | CAATAGATTTTGCTGTGCCATTTT |
| tadG-Probe | FAM-AAAACACAGCCTAAGAAT-NFQ |
|
| |
| ltxD-F | TGTTAAAACTGACCGCACTTGGT |
| ltxD-R | CTTGAGAAAGCGATGTCTTCGTT |
| ltxD-Probe | FAM-CCGAAGCGGATACGTTA-NFQ |
|
| |
| fur-F | TGAAGGCAATAAATCAGTTTTCGA |
| fur-R | CGGTGCAGATAATGTGGTCATG |
| fur-Probe | FAM-TGGCGCCAACCC-NFQ |
|
| |
| Primers for RT-PCR | |
| JA01-F | TTCTACGCAAGGTTGGTTTATTT |
| JA01-R | TGTTATCTACGCACTATATCAAATTCA |
|
| |
| JA02-F | ATTTTGCACAAGATAATGTGTTGG |
| JA02-R | AAATTTGTCCCTTTACACAAATTTTA |
|
| |
| JA03-F | AAAGCCACGTAATAACTGCTGA |
| JA03-R | GAAAACACAGAGTTATTCAAAGGTCA |
|
| |
| JA04-F | TTTTGATTAAAAATTAAGCGTTCG |
| JA04-R | AAGCCTGCGATTTTCCTGTA |
|
| |
| SYBR GREEN Primers for RT-qPCR | |
|
| |
| hemU-F | CGTTATTGATGATGCGTTGG |
| hemU-R | GGAATAATCAAGCCGATCCA |
|
| |
| murD-F | CCTTAACCGGTTTGCAGGTA |
| murD-R | CAATGGCGTGTTCTATGGTG |
|
| |
| rsgA-F | CACACCAATTCGCTTCGTTA |
| rsgA-R | CTTCGTGCTGTTCTTCAACG |
|
| |
| hitC-F | CTTGGGTAACGGCAAAGGTA |
| hitC-R | TTCATCCAGGGCACTAAAGG |
|
| |
| galE-F | CCGTGTTCGGTAGCGACTAT |
| galE-R | TGTCATTTGCCTGCTCAAAG |
|
| |
| glyA-F | GGATTCTCCGCGTATTCTCA |
| glyA-R | CGCCTAAGGTTTTGTGGGTA |
|
| |
| 16S rRNA-F | ACGCGAAGAACCTTACCTACTCT |
| 16S rRNA-R | CCTAAGGCACAAACCACATCTCT |
Underline denotes restriction sites of enzyme listed to the right side of the sequence.
Primer sequences based on HK1651 sequence unless otherwise noted.
TaqMan probe and primer sequences based on D7 sequence.
MGB, minor groove binder; FAM, fluorophore; NFQ, non-fluorescent quencher.
Reverse transcription PCR (RT-PCR)
To synthesize complementary DNA (cDNA), total RNA (5 μg)was pretreated with 1 μl of Turbo DNase (Ambion) and RNA was re-quantified using the NanoDrop spectrophotometer. A 2-μg aliquot of DNase-treated RNA was used as template to synthesize first strand cDNA using random primers as per the manufacturer’s protocol (Invitrogen). Superscript II reverse transcriptase (200 U; Invitrogen) was added to the reaction tubes only. As a negative control for each RNA template, DEPC-treated water was added in place of Superscript II. For RT-PCR experiments, 5 μl of cDNA was used for each reaction. The primers used are indicated in Table 2. Reactions were run as follows: one cycle of 2 min at 95°C, followed by 25–28 cycles of 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C followed by a final cycle of 72 C for 5 min. Samples were resolved on a 1% agarose gel and stained with ethidium bromide. Genomic DNA was used as template for the positive control and no template was used for the negative control.
Real-time quantitative PCR (RT-qPCR)
cDNA was synthesized from total RNA, as described above. TaqMan primers and probes used for RT-qPCR (Primer Express, Version. 2.0; Applied Biosystems) are listed in Table 2. All reactions were run in triplicate according to manufacturer’s protocol, and repeated from three independent biological replicates. Controls included: (1) amplification with a primer/probeset for 16S rRNA, (2) amplification with a reaction mixture without a template, and (3) amplification without the reverse transcriptase. Reaction plates were processed on an Applied Biosystems 7500 real time PCR system as follows: hot-start AmpliTaqGold polymerase was activated at 95°C for 10 min followed by 40 cycles of denaturation for 15 s at 95°C with annealing and extension for 60 s at 60°C. The non-Fe regulated gene glyA was used as a control, and 16S rRNA was used as an endogenous control.
For RT-qPCR analysis using SYBR green dye, specific primers were designed for selected genes, as indicated in the Table 2. Each primer set was pretested by PCR using genomic DNA as template to ensure a single amplicon was produced. For RT-qPCR, each reaction mixture (total volume, 25 μl) contained 5 μl of template, 3.5 μl of DEPC water, 12.5 μl of Power SYBR green PCR master mixture(Applied Biosystems), and 2 μl each of the forward and reverse primers (100 nM). RT-qPCR was performed using Applied Biosystems 7500 Real-time PCR System with the following thermal cycle recommended for the Power SYBR green PCR master mixture: 95°C for 10 min and then 40 cycles of 30 s at 95°C and 1 min at 56°C. Dissociation curves were generated by incubating reaction products at 95°C for 1 min and at 56°C for 30 s and then incrementally increasing the temperature to 95°C. Fluorescence data were collected at the end of the 56°C primer annealing step for 40 amplification cycles and throughout the dissociation curve analysis. Analysis of the melting curves with both primer sets revealed a single sharp peak. The non-Fe regulated gene glyA was used as a control, and 16S rRNA was used as an endogenous control.
Amplification data were analyzed using ABI Primer design SDS 2.1 software (Applied Biosystems). Relative quantification of gene expression was performed by the Ct method, with 16S rRNA expression serving as an endogenous control to normalize target expression within each sample (Winer et al., 1999). By comparing the threshold cycle, the relative expression of a given gene was determined. Relative quantification of gene transcription from JA03 overexpression strain HKR(pJAK16-JA03) was calibrated to HKR.Nal levels. Statistical significance was determined by Student’s t-test.
Ferric uptake regulator titration assay (FURTA)
FURTA, performed as previously described, employs a Fur-regulated lacZ fusion as a reporter gene that allows the detection of transformants carrying multicopy Fur-binding sites as Lac+ colonies on MacConkey agar plates (Stojiljkovic et al., 1994). In brief, potential sRNA promoter sequences were amplified by PCR using the primers listed in Table 2 with genomic DNA (MasterPure DNA Purification kit, Epicentre) from HK1651R as template. Amplicons were cloned into the pGEM-T cloning vector (Promega) according to manufacturer’s instructions, and subsequently transformed into E. coli TOP10 cells. Transformants were selected on LB agar containing ampicillin, and confirmed by colony PCR using the T7 forward primer (T7-F) and the appropriate sRNA-specific reverse primer (Table 2). Plasmid DNA was isolated (Qiagen) and transformed into E. coli H1717, a Fur+ strain containing the Fur-regulated fhuF gene fused to lacZ. To perform the FURTA, transformants were screened for the Lac+ phenotype on Difco™ MacConkey lactose agar plates (Becton, Dickinson and Co.) containing 100 mg ml− ampicillin in the presence or absence of 50 μM FeSO4 supplementation after 30 h of growth at 37°C. In the Fur+ H1717 in the presence of Fe, Fur binds to the fhuF promoter region preventing lacZ reporter gene expression that results in white colonies on MacConkey lactose agar plates. However, Fur will not bind to the fhuF promoter region in the absence of Fe or in the presence of both Fe and Fur-binding sequences carried on a multicopy plasmid that will titrate out the Fur protein from the binding site within the reporter construct. In both of these latter cases, the lacZ reporter gene is expressed and the colonies appear red on MacConkey lactose plates.
β-galactosidase assay
For quantitative analysis of FURTA experiments, E.coli H1717 cells containing the sRNA promoter constructs with putative Fur-box sequences grown on MacConkey plates supplemented with 50 μM FeSO4 were suspended in LB medium (Fe rich, 10–30 μM Fe3+) containing 100 mg ml− ampicillin and grown overnight. The cells were diluted 1:20 in fresh LB-ampicillin medium and grown to stationary phase. Cells were permeabilized and the β-galactosidase activity was measured as previously described (Miller, 1992). Each sample was analyzed in triplicate from two independent assays, and the data pooled for statistical analysis.
Overproduction and purification of Fur
For use in subsequent Fur-binding assays recombinant Fur protein was overproduced using the T7 RNA polymerase/promoter system. The corresponding coding sequence of the fur gene from A. actinomycemcomitans database strain HK1651 was PCR amplified and cloned into the Nco1/Not1 sites of a pET-30b expression vector (Novagen). Novasingles competent E. coli cells (Novagen) were transformed with the fur construct (pAaFur). Transformants were selected on kanamycin-containing LB agar plates and the sequence of the selected clone verified by nucleotide sequencing. The plasmid (pAaFur) was isolated using QIAprep miniprep spin columns (Qiagen) and transformed into E.coli expression host BL21(DE3) strain. Transformants were selected on LB agar containing kanamycin and grown at 37°C. Transformants containing pAaFur were confirmed by PCR using primers (Fur-KpnI-F and Fur-BamHI-R, Table 2) and by nucleic acid sequencing. To induce Fur expression, the selected transformant (BL21(DE3)(pAaFur) was grown in LB broth containing kanamycin to OD600 0.7–0.8, isopropyl-β-D-thiogalactopyranoside(IPTG) (0.5 mM–1 mM) was added, and the culture was shaken 3 h at 37°C or 16 h at 25°C. After induction, the cell lysate was produced by sonication and unbroken cells were removed by centrifugation. The overexpressed recombinant protein was identified on Coomassie-stained 14% SDS-PAGE gels in comparison to whole cell lysates from uninduced cells. Results were confirmed on Western blot using an E. coli anti-Fur antibody obtained from Dr. Michael Vasil (University of Colorado).
Electrophoretic mobility shift assay (EMSA)
Non-radioactive EMSA was performed using the DIG gel shift kit (Roche Applied Science) as described by manufacturer’s instructions. DNA probes specific to the promoter of each sRNA were amplified from chromosomal DNA template by PCR using the primers listed in Table 2. End-labeling of the probes was carried out with digoxigenin-11-ddUTP(DIG-ddUTP) using terminal transferase in 20 μl reaction buffer containing 0.2 M potassium cacodylate, 0.25 MTris-HCl, pH 6.6, 0.25 mg BSA ml−1 and 5 mMCoCl2 incubated at 37°C for 15 min, and then 2 μl0.2 M EDTA (pH 8.0) was added to terminate the reaction. Binding reactions were carried out with 10 nM DIG-labeled probe and cell lysate from pAaFur plasmid (6 μM) in 20 μl reaction buffer containing 20 mM HEPES, pH 7.6, 1 mM EDTA,10 mM NH4)2SO4, 1 mM DTT, Tween 20 (0.2 %, w/v),30 mM KCl, 1 μg poly(dI-dC), and 0.1 μgpoly-L-lysine incubated at 37 °C for 15 min. For the competition reaction, cell lysate was first incubated in presence of 100-fold molar excess of competitor DNA (unlabeled specific DNA) followed(Vogel et al., 2009) by incubation with the labeled probe. E. coli anti-Fur antibody (obtained from Dr. Michael Vasil), diluted 1:50 or 1:250 in PBS, was preincubated with cell lysate prior to incubation with labeled probe for supershift experiments. The bound product was resolved on a 5% polyacrylamide gel in 0.25X TBE buffer at 4°C. The gel was electroblotted onto a nylon membrane and treated with anti-DIG–alkaline phosphatase conjugate. DNA was detected by exposure to autoradiography Hyperfilm ECL.
Engineering the fur mutant
A mutation in the fur gene was constructed by transposon mutagenesis using the suicide vector pMB78 (Bhattacharjee et al., 2007). The fur gene was PCR amplified with primers engineered with KpnI and BamHI restriction enzyme sites (Table 2). The fur gene was cloned into the TA-cloning vector pGEM-T (Promega), transformed into chemically competent E. coli TOP10 cells (Invitrogen) and plasmid was isolated (Qiagen) according to manufacturer’s instructions. Following digestion of plasmid pGEMT-fur with the appropriate restriction enzymes, gel-purified fragments (Qiagen) were ligated into pMB78 to generate pMfur, which was then transformed into E. coli TOP10 cells. Transformants were selected on LB agar supplemented with ampicillin and confirmed by colony PCR using primers specific for the fur gene (Fur-KpnI-F and Fur-BamHI-R, Table 2). In vitro transposon mutagenesis (EZ-Tn5<KAN-2>; Epicentre Biotechnologies) was performed according to manufacturer’s instructions on isolated plasmid to insert a kanamycin resistance (Kmr) cassette into the fur gene. After transformation of E. coli TOP10 with the intact plasmid, transformants were selected on LB agar supplemented with kanamycin and verified by colony PCR using the appropriate primers listed in Table 2. Plasmid was isolated from colonies containing the Kmr cassette in the proper orientation in the fur gene. Plasmid pMfur.KanEZ linearized by digestion with XmaI enzyme was either electroporated or transformed into chemically competent A. actinomycetemcomitans HKR.Nal to generate a fur::kan insertion mutant (Bhattacharjee et al., 2007, Sreenivasan et al., 1991). Transformants were selected on TSBY agar plate containing kanamycin (40 μg/ml). Correct incorporation of the fur::kan insertion mutation sequences into chromosomal DNA was verified by PCR using fur primers Fur-KpnI-F and Fur-BamHI-R, as well as forward primer (ActF-194-F) from the upstream gene actF and reverse primer Km-R from the Kmr cassette, and nucleotide sequencing (Biopolymer Facility of Roswell Park Cancer Institute, Buffalo, NY, USA).
The functionality of the fur::kan mutation was determined using a Fur-deficient E. coli H1780 strain by testing its capacity to complement the Fur deficiency of E. coli H1780. The Fur-deficient E. coli H1780 strain contains a Fur-regulated fiu gene fused to a lacZ reporter gene. The fiu gene is constitutively expressed regardless of the iron conditions due to an engineered fur mutation in this strain. Consequently, β-galactosidase is also constitutively expressed. The A. actinomycetemcomitans fur gene was PCR amplified using specific primers from the parental and HKR.fur::kan strains, and cloned into pGEM-T plasmid generating pGEMT-fur and pGEMT-fur::kan, respectively. These plasmids, as well as the vector only (pGEM-T), were transformed into E.coli H1780. The H1780 strain with pGEM-T alone, H1780 with A. actinomycetemcomitans with wild-type fur, and H1780 with mutated fur were grown in iron-replete LB broth. β-Galactosidase activity was determined from stationary phase cells, as described previously (Miller, 1992), but without IPTG induction.
Genetic complementation of fur mutation
The broad-host-range plasmid pJAK16 was used for fur complementation. The pJAK16 plasmid is an IncQ expression vector that contains a chloramphenicol-resistance gene and an IPTG-inducible Ptac promoter located upstream from a multiple cloning site (Thomson et al., 1999). Plasmid pJAK-fur was constructed by restriction enzyme digestion of pMfur (Table 1) with KpnI/BamHI, yielding a 506 bp fur fragment which was gel purified and ligated into the KpnI/BamHI sites of pJAK16. The pJAK-fur plasmid and pJAK16 vector alone (negative control) were each transformed into chemically competent E. coli SK140, a TOP10 strain carrying RK2 oriT-defective mobilization plasmid pRK21761 (Table 1), as previously described (Thomson et al., 1999), resulting in SK140(pJAK-fur) and SK140(pJAK16), respectively. Each E. coli strain was then used as the donor for conjugation with the HKR. fur::kan mutant strain, using a protocol previously described for rough phenotype strains of A. actinomycetemcomitans (Thomson, et al., 1999). Transconjugants were selected on TSBY agar containing chloramphenicol (2 μg/ml) and naladixic acid (20 μg/ml. Uptake of pJAK-fur was confirmed by colony PCR using specific primers for the fur gene (Fur-KpnI-F and Fur-BamHI-R) and the pJAK-16 plasmid (pTac-F and pJAK16-R), and by nucleotide sequencing. Plasmid-harboring strains were grown in TSBY containing chloramphenicol (2 μg/ml); IPTG induction was not required for gene expression, since the Ptac promoter is leaky.
Overexpression of JA03 sRNA
The JA03 gene was PCR amplified with primers engineered with KpnI and BamHI sites (Table 2), cloned into pGEM-T, and transformed into E. coli TOP10. Following isolation of plasmid pGEMT-JA03 (Qiagen), the JA03 fragment (650 bp) produced by digestion with KpnI/BamHI was gel purified and cloned into pMB78 to yield pMJA03 (Table 1). The sRNA overexpression plasmid (pJAK-JA03) was constructed by enzyme restriction digestion of plasmid pMJA03 with KpnI/BamHI. The resulting JA03 fragment (650 bp) was gel purified and ligated into the KpnI/BamHI sites of the broad-host-range plasmid pJAK16 donated to us by Dr. Jeffrey Kaplan at University of Columbia, NY, which placed JA03 under control of an IPTG-inducible Ptac promoter (Fine et al., 2005). The resulting plasmid, pJAK-JA03, and the pJAK16 vector alone (negative control) were transformed into chemically E. coli SK140. Each E. coli strain, SK140(pJAK16-JA03) and SK140(pJAK16), was used as the donor for conjugation with HKR.Nal, as previously described (Kachlany et al., 2000, Thomson et al., 1999). Transconjugants were selected on TSBY agar containing chloramphenicol and naladixic acid. Uptake of extrachromosomal pJAK-JA03 generating SK140(pJAK16-JA03) was confirmed by colony PCR using specific primers for the gene encoding JA03 (JA03-KpnI-F and JA03-BamHI-R, Table 2), as well as by nucleic acid sequencing. Similarly uptake of the pJAK16 plasmid generating SK140(pJAK16) was confirmed by colony PCR using primers pTac-F and pJAK16-R (Table 2). Plasmid-harboring strains were grown in TSBY containing chloramphenicol (2 μg/ml); IPTG induction was not required for gene expression, since the Ptac promoter is leaky.
Microtiter plate biofilm assay
Biofilm growth and quantification was determined using a modification of a standard microtiter plate biofilm assay, as described previously (Haase et al., 2006). Briefly, frozen stock cultures were plated onto TSBY agar and incubated in a candle jar for 72 h at 37°C. One to five isolated colonies were transferred to TSBY broth and grown overnight. Cultures were standardized to OD600 of 0.050 and cells were harvested by centrifugation and resuspended in TSBY, and applied to 96-well, flat-bottomed, untreated polystyrene microtiter plates (Nalge Nunc International, Rochester, NY, USA). Plates were covered and incubated for 21 h or 48 h in 5% CO2 at 37°C. Growth was monitored at OD595 using a microplate reader (Beckman Coulter AD340). Culture supernatants were decanted and unbound bacteria were removed by washing with phosphate buffered saline, pH 7.2. Biofilm cells were fixed with methanol and air-dried prior to staining with 0.1% (w/v) crystal violet (Sigma, St. Louis, MO, USA) for 5 min. Dye was decanted and wells washed with distilled water until negative control wells were clear. After air-drying the plates, ethanol was used to solubilize the bound crystal violet. Plates were mixed briefly and the absorbance of the dye quantified at 595 nm using a microplate reader. Each sample was tested in three biological replicates with six replicates per assay. Statistical significance was determined by the Student’s t-test.
RESULTS
Identification of sRNA in A. actinomycetemcomitans
We identified several Fur-regulated sRNA molecules in A. actinomycetemcomitans HK1651 using a bioinformatics approach based on criteria previously reported to identify sRNA sequences in other bacterial species. The sRNA target prediction tool TargetRNA was then used to identify potential mRNA targets of the sRNA molecules. The region proximal to the ribosome-binding site of each message in the A. actinomycetemcomitans genome was evaluated for a likelihood of interacting with sRNAs by homologous base pairing. TargetRNA predicted statistically significant interactions of four sRNAs (JA01, JA02, JA03, and JA04) with biofilm-associated genes, including the flp fimbrial operon and genes associated with EPS, LPS, and several known Fe-regulated proteins (supplementary information, Table S1) (Balashova et al., 2006b, Carrondo, 2003, Grifantini et al., 2003, Chen et al., 2002). Predicted base-pairing regions of each sRNA with putative targets are shown in supplementary information, Table S2. The putative nucleotide coordinates of each sRNA within the IGRs annotated from the published A. actinomycetemcomitans HK1651 sequence (Roe et al.) are as follows: JA01 (1366985–1367137 within IGR1299), JA02 (1109159–1109274 within IGR1069), JA03 (1446694–1446967 within IGR1375), and JA04 (356690–356770 within IGR0328). Computer modeling using Mfold predicted that each of these sRNAs had a stable RNA secondary structure (supporting information, Fig. S1).
Transcription of sRNAs
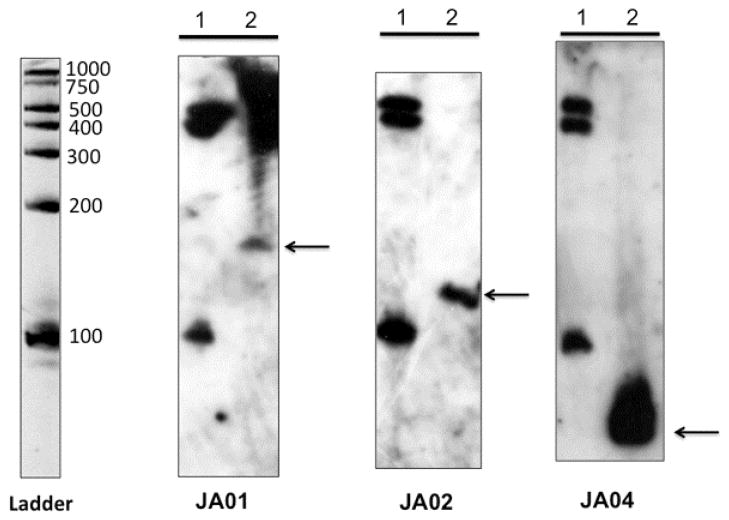
To verify that each candidate sRNA is indeed transcribed as a relatively small transcript and is not part of the downstream transcript, Northern blot analysis was performed. Total RNA was isolated from early stationary phase, separated on denaturing acrylamide gels, transferred to Hybond N+ membranes, and probed with biotinylated DNA probes corresponding the complementary DNA sequence of each sRNA. The PCR amplicon used to generate the dsDNA probe was included on the blot as a positive control (data not shown). Transcript sizes were estimated from the RNA Century Plus Ladder (Ambion); the 500, 400, and 100 base markers consistently were detected on the blot using the Bright-Star Psoralen-Biotin kit, regardless of the probe. Bands visible for JA01 (ca. 155 nt), JA02 (ca. 105 nt), and JA04 (ca. 76 nt), as shown in Fig. 1, correlated well with the predicted size for JA01 (153 nt), JA02 (116 nt), and JA04 (81 nt) (supporting information, Fig. S1). Several attempts were made to obtain a transcript for JA03 on Northern blot using both agarose gels and different percentage acrylamide gels. Also, dsDNA probes from different regions in the putative sRNA region were used. The dsDNA probe could detect the probe loaded onto the gel, but no transcripts could be detected for JA03. This could be due to secondary structure problems with the RNA on the gel and/or lower sensitivity of DNA probes (Vogel et al., 2009).
Fig. 1.
Northern blot analysis of sRNA. The RNA Century Plus size ladder was loaded in lane 1 of each gel. The ladder was visualized by ethidium bromide staining as shown (inverted photograph). Total RNA was extracted from parental strain HKR.Nal grown in TSBY with 300 μM dipyridyl chelator. About 10 μg of RNA was loaded in lane 2. After electrophoresis each gel was electroblotted onto positively charged nylon membranes and hybridized with biotinylated dsDNA probes specific to JA01, JA02, and JA04, as indicated. Hybridization was detected with the BrightStar Psoralen-Biotin kit.
Potential coding capacities of the sRNAs
The nucleotide sequence of each sRNA was examined for potential open reading frames in all three reading frames using DNASIS (ver. 3.0). There were no potential peptides greater than 2,668 Da for JA01, 1,303 Da for JA02, and 3,096 Da for JA04. Since the size of the transcript for JA03 has yet to be determined, the entire IGR potentially encodes peptides no greater than 1,809 Da, while the putative JA03 associated with a promoter containing a Fur box encodes for peptides no greater than 1,614 Da.
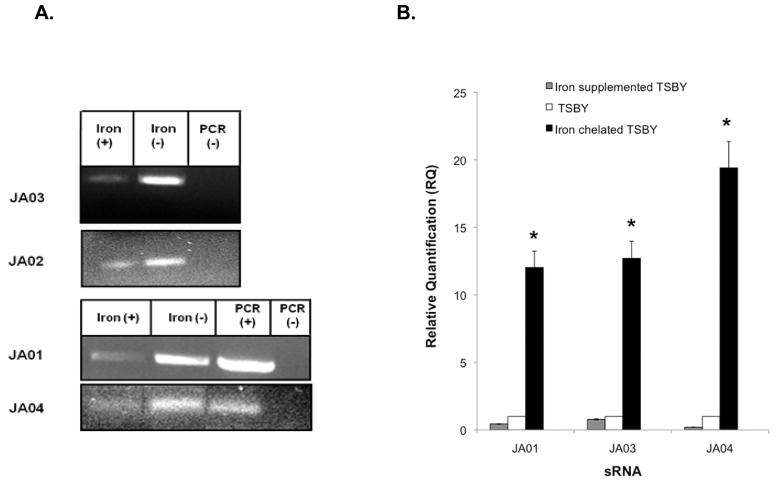
Fe-responsive transcription of sRNAs. Initially, to determine if Fe limitation had any effect on sRNA transcription, reverse transcription PCR (RT-PCR) was done comparing cells grown in Fe-sufficient conditions (TSBY) with Fe-chelated TSBY. Each of the sRNA molecules (JA01-JA04) was expressed to a greater extent under Fe-limited (chelated) growth conditions (Fig. 2A). Then to more accurately quantify the difference between these conditions, and to determine if additional Fe had any effect on transcription, two-step real-time PCR (RT-qPCR) was performed with the addition of Fe-supplementation of TSBY to the assay. Transcription of JA01, JA03 and JA04 increased by 12-, ~12- and 19-fold, respectively, in Fe-chelated conditions when compared to growth in TSBY (Fig. 2B). Fe-supplemented TSBY suppressed sRNA transcription relative to TSBY. Due to the short length and the nucleotide sequence of the IGR (211 bp) containing JA02, a reliable TaqMan probe could not be designed for the RT-qPCR uniform cycling conditions.
Fig. 2.
Fe regulation of sRNA candidates. (A) RT-PCR analysis: comparison of sRNA transcript expression in HKR.Nal grown in TSBY (Fe sufficient) and Fe-chelated TSBY, genomic DNA (PCR + control), and no template (PCR–control). (B) RT-qPCR analysis using TaqMan primers and probes: quantitative expression sRNA molecules JA01, JA03, and JA04 under Fe-supplemented TSBY, TSBY (Fe sufficient), and Fe-chelated TSBY growth conditions normalized to 16S rRNA gene expression. Expression level of each transcript under the Fe-supplemented or Fe-chelated conditions was relative to the expression in TSBY. * indicates a significant difference (P < 0.01) of the test condition from the control.
Fur regulation of sRNAs
In many cases, genes that respond to Fe are commonly regulated by Fur, a global Fe-dependent transcriptional regulator that binds to operators (Fur boxes) located proximally to the promoters (Bosch et al., 2002, Kirby et al., 2001, Najimi et al., 2008, Osorio et al., 2004). In silico analysis determined that each sRNA possessed a Fur box-like motif proximal to the putative −35 and −10 promoter sequences. These Fur box-like motifs were 63–74% (12–14 of the 19 bp) identical to the consensus Fur box of E. coli (supporting information, Table S3). Previously, a Fur box-like sequence with 68% identity to the consensus Fur box of E. coli was located downstream of a −35 and −10 sequence in the fur gene of A. actinomycetemcomitans (Haraszthy et al., 2002). The Fur box of the fur gene of A. actinomycetemcomitans is 52–65% identical to the putative sRNA Fur box motifs reported in this study.
While a consensus 19-bp nucleotide sequence for the Fur box has been determined for E. coli, it is clear that Fur binds to Fur boxes having only partial homology to that consensus sequence (Bosch et al., 2002, Kirby et al., 2001, Najimi et al., 2008, Osorio et al., 2004). FURTA (Fe uptake regulator titration assay), a genetic assay for detecting Fur boxes, is routinely used to evaluate the Fur-binding potential of DNA sequences from various bacterial species that respond to Fe (Bosch et al., 2002, Kirby et al., 2001, Najimi et al., 2008, Osorio et al., 2004). Genes from other bacteria possessing putative Fur boxes that differ from the consensus sequence (e.g., Bordetella avium (13/19 bp), Photobacterium damselae ssp (14/19 bp), Aeromonas salmonicida (13/19 bp) and Pasteurella multocida) are strongly positive in FURTA (Bosch et al., 2002, Kirby et al., 2001, Najimi et al., 2008, Osorio et al., 2004). Thus, FURTA was used to determine whether the putative Fur-box nucleotide sequences of each sRNA had the capacity to bind Fur.
Colonies of E. coli strain H1717 transformed with the control plasmid pGEM-T alone exhibited only a minimal LacZ expression phenotype (pale pink) when plated on Fe-supplemented MacConkey agar. In contrast, colonies of strain H1717 transformed with plasmids encoding each of the four sRNA promoter constructs (pGEMT-JA01pro, pGEMT-JA02pro, pGEMT-JA03pro, and pGEMT-JA04pro) exhibited dark red colony morphologies (Table 3). Quantitation of β-galactosidase activity by the transformants confirmed the FURTA results (Table 3). These data strongly indicated that each of the DNA fragments encoding the sRNA putative promoter constructs was capable of titrating Fur in an iron-replete environment.
Table 3.
FURTA analysis of the sRNA promoters.
| sRNA promoter plasmid | Lac-FURTA1 | Mean β-galactosidase activity (±SD)2 |
|---|---|---|
| pGEMT-JA01pro | + | 603 (±85) |
| pGEMT-JA02pro | + | 533 (±22) |
| pGEMT-JA03pro | + | 426 (±46) |
| pGEMT-JA04pro | + | 681 (±18) |
| pGEMT-alone | − | 176 (±26) |
Lac phenotype of the E.coli H1717 (Fur+) containing the indicated plasmid after 30 h incubation, 37°C on MacConkey agar supplemented with 50 μM FeSO4. Lac+ (red), lac− (pale pink).
β-galactosidase activity, in Miller units, of E.coli H1717 containing the indicated plasmid grown to stationary phase in LB medium. SD, standard deviation.
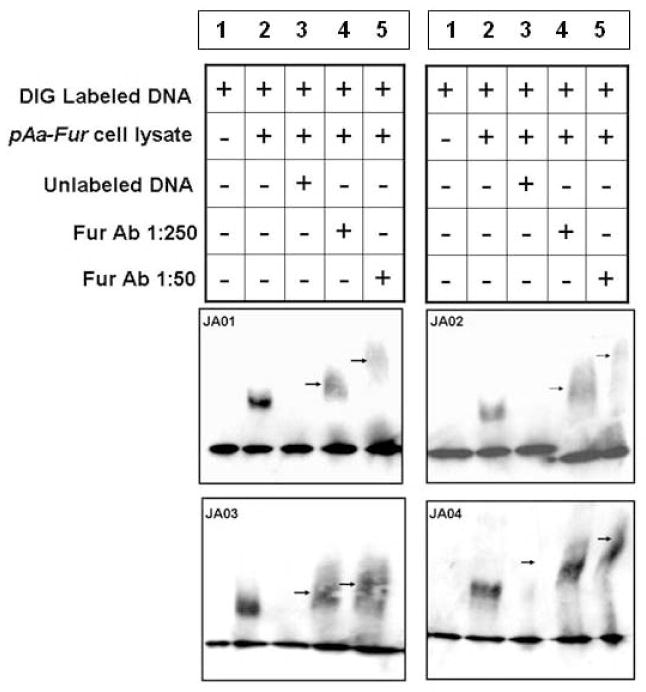
To better define the specificity of Fur binding to the predicted Fur box sequence of the four sRNA molecules, electrophoretic mobility shift assays (EMSA) were performed using cell lysates from E. coli overexpressing the A. actinomycetemcomitans Fur protein (AaFur) (Fig. 3). As expected, no band shift in mobility was observed for each DIG-labeled sRNA promoter DNA in the absence of cell lysate (Fig. 3, lane 1). Preincubation of each DIG-labeled sRNA promoter DNA with 0.4 μg of cell lysate of E. coli overexpressing the AaFur, however, was associated with a shift in fragment mobility resulting in a band shift (Fig. 3, lane 2).
Fig. 3.
Electrophoretic mobility shift assay (EMSA). To demonstrate the binding of Fur to predicted sRNA promoters, digoxigenin-labeled DNA probes corresponding to JA01, JA02, JA03, and JA04 sRNA promoter regions were used in EMSAs with cell lysates from a plasmid overexpressing AaFur protein (pAaFur). Mouse anti-Fur antibody (diluted 1:50 and 1:250) was used to show the supershift of DNA-protein-Ab complex (arrows).
To ensure that AaFur and not EcoliFur caused the decreased mobility of the promoter sequence, E. coli lysate without AaFur failed to show a shift (data not shown). Unlike the FURTA, in which the titration of relatively small amounts E. coli Fur by the Aa Fur boxes transiently frees enough of the reporter gene of Fur repression to produce indicator enzyme, the gel shift assay requires relatively high levels of the Fur repressor to bind tightly and constantly to produce a shift in mobility. Only E. coli overexpressing AaFur was able to cause a gel shift. To confirm the association between AaFur and the Aa Fur box, a competition reaction was performed in which a 100-fold molar excess of unlabeled, sRNA-specific promoter DNA was employed. Shifting of mobility of the sRNA fragments in EMSA was inhibited by preincubation with this excess of unlabeled probe (Fig. 3, lane 3).
When anti-E.coli Fur antibody was added to the reaction, a supershifted band appeared in the gels (Fig. 3, arrows, lanes 4, 5). The mobility shift correlated with antibody concentration, i.e. more antibody (1:50 dilution) produced a greater shift than less antibody (1:250 dilution). The binding of anti-Fur antibody to the DNA:Fur complex further retarded the mobility, and the more antibody bound the greater the retardation of mobility. These data strongly indicate that the mobility shifts observed in the EMSA were due to the direct interaction between AaFur and the Fur boxes located in the DNA fragments encoding the sRNA promoters.
Characterization of a fur mutant
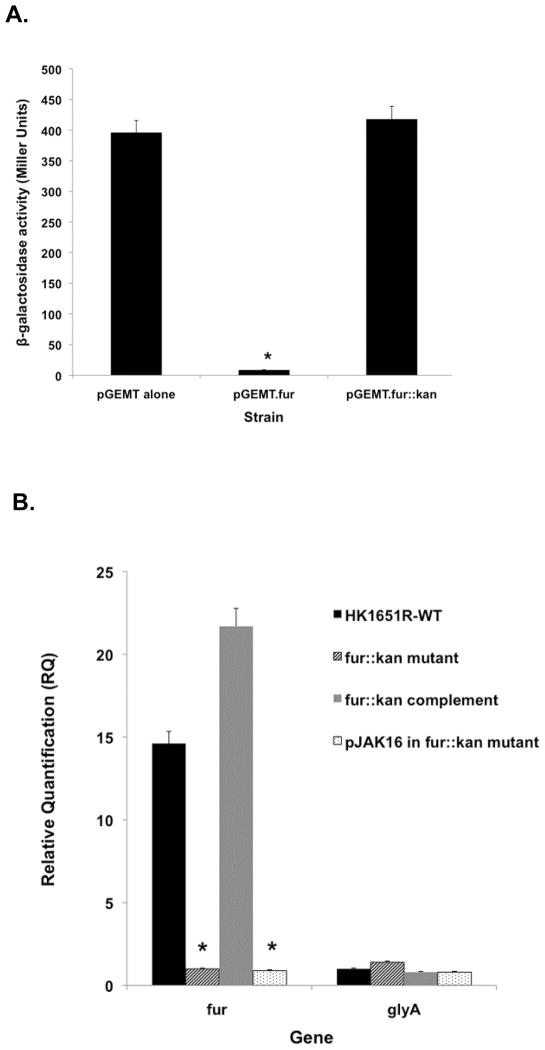
To further investigate the role of the fur gene in regulating sRNAs in A. actinomycetemcomitans, a fur mutant strain was constructed by inserting a Kmr cassette into the fur-encoding region of the parental strain via transposon mutagenesis. The lack of functionality of the fur::kan mutation was verified by testing its capacity to complement the Fur deficiency of E. coli strain H1780 relative to HK1651R Fur. As expected, introduction of functional A. actinomycetemcomitans Fur on plasmid pGEMT.fur into E. coli H1780 was able to restore the defect of the E. coli H1780 strain and shutdown β-galactosidase production. The mutated fur gene carried on plasmid pGEMT.fur::kan was unable to provide a functional Fur, and unable to complement the Fur defect of the H1780 strain; thus, constitutive β-galactosidase activity was observed in H1780, similar to pGEM-T vector alone (Fig. 4A). These data confirm that the Kmr cassette successfully disrupted the fur gene in the parental strain rendering it nonfunctional.
Fig. 4.
(A) Fur-complementation assay. Fur-deficient E. coli H1780 was used to test for the ability of plasmids to complement the Fur deficit. The promoter of the Fur-regulated fiu gene is fused to a promoterless lacZ. β-galactosidase is constitutively expressed in H1780. Samples: pGEMT-(vector alone); pGEMT.fur (strain complemented with Aafur); pGEMT.fur::kan (Aafur disrupted with Kmr cassette). (B) Effect of the fur::kan mutation and fur mutation complementation on the expression of fur. RT-qPCR was used to quantify fur expression in parental HKR.Nal and fur::kan mutant strains. The non-iron regulated glyA gene was used as a control. 16S rRNA was used as the endogenous control. * indicates a significant difference (P < 0.05) of the test condition from the control.
The transcript levels of fur in HKR.fur::kan and in the parental strain HKR.Nal were compared by RT-qPCR. As expected, the fur gene was transcribed ~15 fold (P = 0.02) more in the parental strain than in the fur::kan mutant strain (Fig. 4B). Introducing a functional copy of fur carried on the pJAK-fur expression plasmid restored the fur deficiency of HKR.fur::kan. The complemented fur::kan strain, HKR(fur::kan-pJAKfur), had a 21.6-fold (P = 0.008) increase in fur transcription compared to mutant strain HKR.fur::kan, while fur expression in the fur::kan mutant strain with pJAK16 vector alone (HKR.fur::kan-pJAK16) was similar to that of HKR.fur::kan. RT-qPCR data, together with quantitative FURTA experiments confirm that fur gene expression was successfully interrupted by insertion of the Kmr cassette and that the fur::kan strain likely produced a non-functional or partially functional Fur.
Effect of Fur on transcription of sRNAs
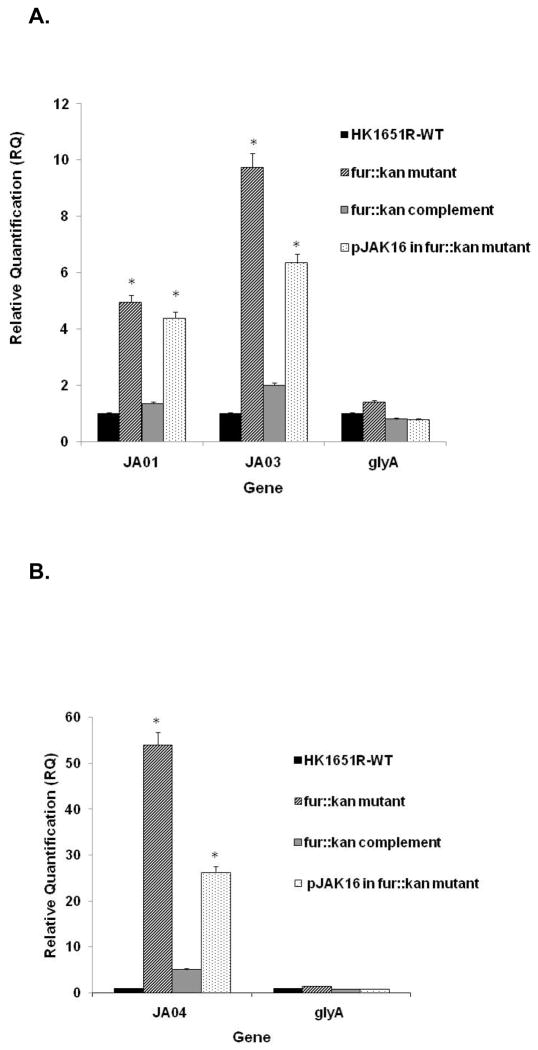
To determine the role of Fur on the expression of selected sRNA molecules (JA01, JA03 and JA04), the transcript levels of these genes was examined using RT-qPCR comparing the fur::kan mutant strain and parental strain grown in TSBY (Fe-sufficient) medium. Interestingly, the expression of all three sRNA molecules was significantly increased in the fur::kan mutant strain compared to the parental strain (Fig. 5A,B); notably, expression of JA01 increased (4.9 fold, P = 0.032), JA03 (9.7 fold, P = 0.039) and JA04 (53.9 fold, P = 0.007). When the fur::kan mutant strain was complemented by introduction of fur on an overexpressing plasmid, transcription of these sRNA molecules was reduced to the level in the parental strain. The sRNA expression level in the pJAK16 vector control strain was similar to the fur::kan mutant strain (Fig. 5A,B). These data indicate that these sRNA molecules are under direct control by the Fur master regulator.
Fig. 5.
Effect of fur::kan mutation on expression of sRNAs. Expression of sRNA molecules JA01, JA03, and JA04 in parental HKR.Nal (indicated as HK1651R) and fur::kan mutant strain in RT-qPCR. The non-iron regulated glyA gene was used as control. 16S rRNA was used as an endogenous control. * indicates a significant difference (P < 0.05) of the test condition from the control.
The possibility of polar effects of fur::kan mutation on upstream and downstream genes was evaluated using RT-qPCR. Transcription of the flavodoxin-encoding gene, fldA, located upstream of fur, and AA02518 a gene directly downstream of fur were increased 3.0 fold (P = 0.03) and 2.7 fold (P = 0.09), respectively, in the fur::kan mutant strain as compared to the parental strain. Both fldA and AA02518 are transcribed in the same direction as fur. The fldA gene is known to be iron repressible, and a similar increase in fldA gene transcription in fur mutant strains has been observed in other bacteria (Achenbach & Genova, 1997, Bender et al., 2007, Ghassemian & Straus, 1996). Therefore, it is possible that the expression fldA and AA02518 of A. actinomycetemcomitans are also repressed by Fur, as in other bacteria, which accounts for the increase of transcription of these genes observed in the fur::kan mutant strain.
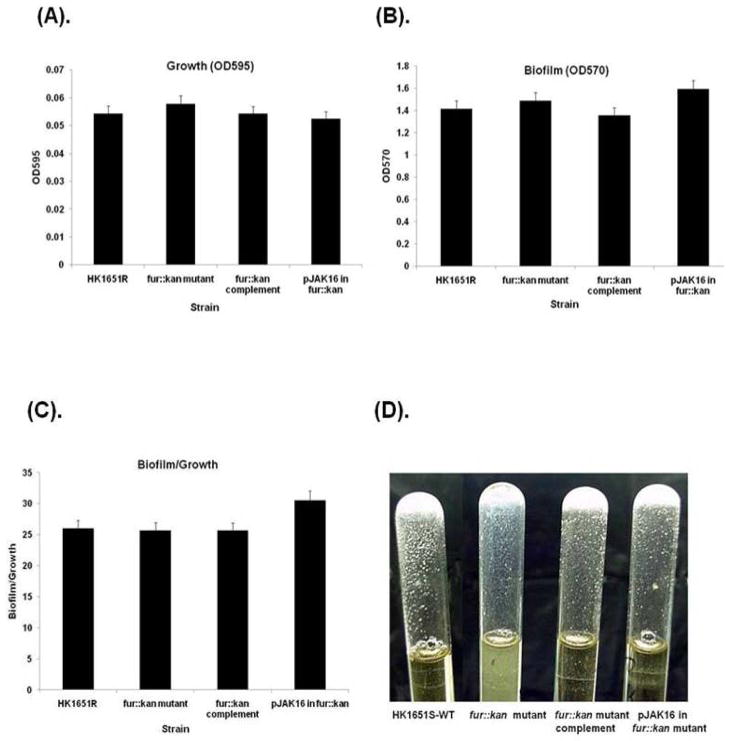
Effect of Fur on biofilm
In order to determine whether biofilm formation is affected by fur mutation, a quantitative biofilm assay was performed in TSBY broth on stationary phase (21 h) cells of the parental, fur::kan mutant, and fur::kan complement strains, as well as the pJAK16 vector alone in the fur::kan mutant strain. The growth and quantity of biofilm mass was not significantly different for any of the strains tested (Fig. 6A,B). When biofilm was standardized to growth, no significant differences were noted among the strains (Fig. 6C). However, when the strains were grown in glass tubes in TSBY broth the fur::kan mutant produced a biofilm with more slime and also produced a slightly turbid broth compared to other strains, as shown in Fig. 6D, suggesting a change in expression of some biofilm determinant genes when grown under these conditions.
Fig. 6.
Effect of Fur on biofilm formation in TSBY. Microtiter plate biofilm assay; (A) Growth of cells as measured by optical density at 595 nm, (B) biofilm mass as measured by absorbance at 570 nm of crystal violet eluted from biofilm cells, and (C) biofilm mass normalized to cell density, i.e. biofilm/growth ratio. (D) Cultures grown 24 h in glass tubes; biofilm formation on surface of the tube and supernatant turbidity. Antibiotic resistant HKR.Nal (designated here as HK1651R) was used as WT.
Effect of JA03 sRNA overexpression on its mRNA targets and biofilm determinant genes
A strain overexpressing JA03 sRNA, HKR(pJAK-JA03), was engineered to determine the regulatory role exerted by JA03 sRNA on cellular functions. The JA03 sRNA insert in the expression plasmid was derived using primers encompassing nearly the entire IGR. In these experiments, JA03 was expressed from a plasmid under the control of the IPTG-inducible Ptac promoter in the adherent variant nalidixic acid resistant clinical isolate HKR.Nal. Thus, JA03 sRNA expression was independent of the Fur repressor, allowing analysis of target behavior independent of growth limitations resulting from Fe starvation stress or Fe toxicity, and in the presence of a functional Fur repressor protein. The positive effect of JA03 overexpression on JA03 sRNA transcription was clearly evident as indicated by RT-qPCR where expression of JA03 sRNA increased ~70 fold (P < 0.01) compared to the parental strain or to HKR(pJAK16) harboring the pJAK16 vector alone. Expression of a control gene, glyA (serine hydroxymethyltransferase), a housekeeping gene with no Fe-binding function, was similarly expressed in both JA03 sRNA overexpression and parental strains.
To examine the effect of JA03 sRNA overexpression on cellular functions, six targets of JA03 sRNA (murD, rsgA, galE, ltxD tadG, and hitC) predicted by TargetRNA and three biofilm determinant genes (tadV, rmlB, pgaC) that had been previously characterized (Amarasinghe et al., 2009) were selected for evaluation by RT-qPCR. The expression of each potential target in HKR(pJAK16-JA03) was compared to the parental strain (Table 4). Interestingly, expression of JA03 from a heterologous promoter resulted in significant repression of transcription of hitC, an ATPase and a member of the hitABC gene cluster coding for a known periplasmic-binding protein-dependent transport system in A. actinomycetemcomitans (Rhodes et al., 2007). JA03 sRNA overexpression correlated with repression of several other genes with non-Fe functions, e.g. murD, a gene that encodes UDP-N-acetylmuramoylalanine-D-glutamate ligase, which is involved in the peptidoglycan-based cell wall biogenesis pathway, and ltxD, a gene encoding a protein involved in leukotoxin secretion. The galE gene that encodes an UDP-glucose-4-epimerase involved in nucleotide sugar metabolic pathway was nominally increased suggesting JA03 sRNA likely does not control its expression. Biofilm-associated genes, tadV and tadG genes of the flp fimbrial operon, LPS-associated rmlB, and EPS-associated pgaC were repressed approximately 1.7–2.1 fold (Table 4) suggesting JA03 sRNA may have a role in the regulation of biofilm formation/adherence in this organism. The non-heme ferritin gene rsgA, another putative target, was also slightly up-regulated. Other potential targets (supplementary information, Table S1) involved in biofilm formation, such as autoaggregation and type IV secretion genes, have yet to be tested by RT-qPCR. Transcriptome analysis of the overexpression strain will give a more global view of genes affected by this sRNA.
Table 4.
Effect of JA03 overexpression on selected genes in rough variant HK1651R by RT-qPCR.
| Gene Accession Number | Gene | Gene Function/Category | Expression of HKR(pJAK16-JA03) relative to HKR.Nal1 | Expression of HKR(pJAK16) relative to HKR.Nal1 |
|---|---|---|---|---|
|
| ||||
| Fold Change (± standard deviation) | ||||
| Up-regulated genes | ||||
|
| ||||
| AA02121 | rsgA | non-heme ferritin | 1.65 ± 0.39 | 1.21 ± 0.30 |
| AA01886 | galE | UDP-glucose-4-epimerase | 1.31 ± 0.1 | 1.09 ± 1.23 |
|
| ||||
| Down-regulated genes | ||||
|
| ||||
| AA00869 | tadV | tight adherence protein V; prepilin peptidase | 1.75 ± 1.1 | ND |
| AA00880 | tadG | tight adherence protein G | 1.71 ± 0.19 | 1.342 |
| AA00492 | pgaC | N-glycosyltransferase | 1.79 + 0.8 | ND |
| AA02623 | rmlB | dTDP-D-glucose 4,6-dehydratase | 2.06 ± 0.1* | ND |
| AA00838 | murD | UDP-N-acetylmuramate-alanine ligase | 3.58 ± 1.1* | 1.012 |
| AA02803 | ltxD | leukotoxin secretion protein D | 3.13 ± 1.2* | ND |
| AA01051 | hitC | Fe (III) ABC transporter, ATP-binding protein | 196 ± 232* | 0.96 ± 0.26 |
ND, not done
HKR.Nal (nalidixic acid resistant HK1651R, parental strain).
Sample assayed once.
Significant threshold: ≥ twofold change in expression with a P value of ≤ 0.05.
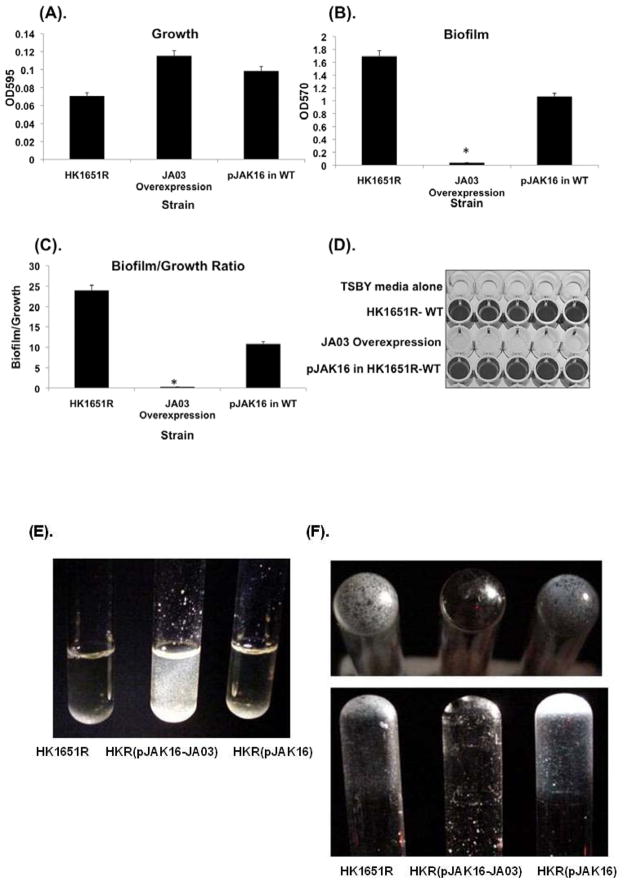
Effect of JA03 sRNA overexpression strain on biofilm formation
To determine if JA03 sRNA overexpression regulates in vitro biofilm formation by A. actinomycetemcomitans, the ability to form biofilm by HKR(pJAK16-JA03) was compared with HKR.Nal. Biofilm growth and quantification was determined using a modification of a standard microtiter plate biofilm plate assay, as described previously. All assays were performed in Fe-sufficient TSBY broth. Although there were no significant differences in growth between the strains after 48 h as measured by OD600, the HKR(pJAK16-JA03) overexpression strain produced less biofilm in comparison to the amount of biofilm produced by either HKR.Nal or HKR(pJAK16) carrying the pJAK16 expression vector alone (Figs. 7A–C). HKR.Nal formed approximately 24 fold (P < 0.05) more biofilm in an untreated, polystyrene microtiter plate compared to the HKR(pJAK16-JA03) overexpressing strain (Fig. 7D). When standardized cultures were grown in glass test tubes for 48 h at 37°C in a candle jar, the JA03 overexpression strain formed large flocs (microcolonies) that clearly did not adhere to the glass surface, unlike the HKR.Nal and vector control strains (Figs. 7E, F). These data demonstrate that JA03 sRNA overexpression did not interrupt intercellular adhesion, but significantly reduced cell adhesion to an abiotic surface.
Fig. 7.
Effect JA03 sRNA overexpression on biofilm formation in TSBY. Microtiter plate biofilm assay: (A) Growth of cells as measured by turbidity at optical density 595 nm, (B) biofilm mass as measured by absorbance at 570 nm, (C) biofilm mass normalized to cell density, i.e. ratio: O.D. 570 nm biofilm/O.D. 595 nm growth, and (D) microtiter plate biofilm stained with crystal violet. Cultures grown 24 h in glass tubes: (E) supernatant after vortexing, and (F) biofilm formation on surface of the tube. * indicates a significant difference (P < 0.05) of the test condition from the control. Antibiotic resistant HKR.Nal (designated here as HK1651R) was used as WT.
DISCUSSION
A bioinformatics screen of the A. actinomycetemcomitans genome identified several potentially Fur-regulated sRNA molecules within intergenic regions. Each of the four sRNA molecules selected for further study, JA01-JA04, possessed a Fur box-like sequence within a probable −35 and −10 promoter sequence that was able to titrate E. coli Fur in a FURTA. This was confirmed by EMSA using E. coli cell lysates overexpressing AaFur, which clearly demonstrated that Fur is able to bind to each sRNA promoter. The transcription of each sRNA molecule in A. actinomycetemcomitans was decreased by Fe supplementation and increased by limiting Fe by chelation demonstrating the influence of environmental Fe availability on gene expression.
This study provides the first evidence of Fe-regulated, Fur-responsive sRNAs in A. actinomycetemcomitans. There are several advantages for bacteria to use sRNA to regulate gene expression. First, their rapid synthesis requires considerably less energy compared to the synthesis of protein regulators. Therefore, the use of sRNA may be more economical to the bacteria under stressful environments (Guillier et al., 2006). Second, sRNA provides bacteria a means to fine-tune the expression of genes, stabilize mRNA, and initiate the expression of colonization factors under Fe-limited conditions. sRNA also provides a rapid means to turn off particular genes when the environment stress signal disappears (Guillier et al., 2006).
Based on in silico analysis using TargetRNA, it is interesting to note that the mRNA of several biofilm determinant genes recently found to be iron regulated (fimbriae, EPS, LPS and luxS) (Amarasinghe et al., 2009, Fong et al., 2003), potentially base pair with the four Fur-regulated sRNA molecules identified in this study. Genes associated with these biofilm determinants do not possess Fur-binding boxes in their promoter regions, suggesting a possible indirect Fur regulatory mechanism.
Regulation of biofilm formation in A. actinomycetemcomitans could be accomplished by several sRNA molecules as in other bacteria where multiple redundant sRNAs act together in response to environmental stress signals to better adapt to their environment (Svenningsen et al., 2009, Tu & Bassler, 2007, Tu et al., 2010). Expression of sRNA molecules under stress conditions and their repression upon disappearance of the particular stress signal is precisely controlled by negative feedback loops that help to fine-tune sRNA-mediated expression of genes in response to the environment (Tu et al., 2010). This type of precise control could also be present in A. actinomycetemcomitans controlling the sRNA molecules.
When JA03 sRNA was overexpressed, multiple Fe- and Fur-regulated genes were repressed, including the hitC gene involved in hitABC Fe-uptake system in A. actinomycetemcomitans. In the mRNA target predictions of JA03 sRNA, it was observed that significant base pairing occurred between JA03 sRNA and hitC mRNA. Previous studies have shown that isogenic mutants of A. actinomycetemcomitans with inactivation of hitA, hitB, and hitC were unable to grow in Fe-limited growth medium compared to the parental strain indicating an essential role of hitABC in Fe acquisition under Fe-stressed conditions (Rhodes et al., 2005). There are two possibilities for sRNA-mediated repression of the hitC gene. First, it is possible that JA03 sRNA binding to the 5′UTR of hitC gene masks the ribosome-binding site to inhibit ribosome entryonto mRNA, as has been described for other bacteria (Sharma et al., 2007, Storz et al., 2004). This blockage of ribosomal entry onto mRNA by sRNA-mRNA pairing at 5′UTR will lead to rapid translational inhibition of these mRNA targets by accelerating the RNaseE-mediated mRNA degradation (Deana & Belasco, 2005, Masse et al., 2003, Morita et al., 2005). In A. actinomycetemcomitans, the hitA (afuA) gene is known to be Fe- and Fur-regulated (Rhodes et al., 2007, Willemsen et al., 1997). Thus, as part of the hitABC locus, repression of hitC transcription likely occurs under iron-sufficient conditions when activated Fur binds to Fe-regulated gene promoters. Therefore, the second reason why hitC may be repressed could be that JA03 sRNA overexpression, as it occurs in an iron-limited environment, may lead to the degradation of iron-containing proteins (i.e. iron storage proteins such as bacterioferritins). This phenomenon, known as the iron sparing effect (Gaballa et al., 2008, Masse & Gottesman, 2002), causes an increase in intracellular Fe concentration, which consequently activates Fur and represses Fur-regulated genes. Similarly, JA03 sRNA overexpression in this study led to the down-regulation of multiple Fe- and Fur-regulated genes, suggesting an occurrence of an iron sparing effect.
Another target of JA03 sRNA was ltxD, part of the ltxCABD operon that is responsible for production of LtxA (leukotoxin). The ltxC gene encodes for a protein responsible for acylation of the toxin, ltxA encodes the toxin, and ltxB and ltxD encode for predicted components of the membrane transport system. LtxA expression is partly regulated by LuxS (also regulated by Fe), and levels of LtxA are affected by cyclic AMP concentration (Fong et al., 2001, Inoue et al., 2001). A. actinomycetemcomitans leukotoxin can also act as a hemolysin that has the ability to lyse erythrocytes resulting in beta-hemolysis of blood agar. This leukotoxin-dependent hemolysis is Fe dependent (Balashova et al., 2006a, Balashova et al., 2006b); hemolysis is suppressed in the presence of FeCl3, while strong hemolysis was observed upon Fe chelation. Since erythrocyte lysis is one mechanism by which a bacterial pathogen acquires Fe, it is suggested that leukotoxin may play a role in Feacquisition (Balashova et al., 2006a, Balashova et al., 2006b).
Down-regulation of the ltxD gene upon JA03 overexpression could be either a direct or indirect effect. JA03 sRNA could directly regulate the ltxD gene at the post-transcriptional level in order to achieve maximum level of efficiency in iron utilization in a low iron environment. One possibility is that JA03 sRNA is degrading the transcripts of nonessential iron-utilizing proteins (i.e. LtxD in order to spare available iron for essential proteins. Alternatively, our studies indicated that expression of ltxD in fur mutant was up-regulated 2.2 fold compared to the wild-type strain indicating that Fur protein is negatively regulating the expression of ltxD gene in wild-type strain in Fe-replete conditions. JA03 overexpression may indirectly down-regulate several iron-regulated genes via an iron sparing effect thereby activating the Fur cascade and down-regulating the ltxD gene.
Expression of biofilm determinant genes tadV (fimbriae), pgaC (EPS), and rmlB (LPS) were only modestly changed by the overexpression of JA03 sRNA, though bacterial cells produced non-adherent flocculation in broth, and little or no biofilm on untreated polystyrene and glass surfaces. This is characteristic of an intermediate phenotype where intercellular adhesion is maintained permitting flocculation, but adhesion to surfaces is abolished. These data also suggest that biofilm regulation goes beyond the regulation of the standard biofilm determinants, yet to be explored through transcriptional profiling of this overexpression strain.
In summary, we have so far identified and characterized at least one A. actinomycetemcomitans sRNA that is Fe-responsive, regulated by Fur, and has an effect on biofilm formation. Although these sRNA appear to base pair with the mRNA of previously identified biofilm determinant genes, their effect on global transcription needs to be further investigated. Due to the multifactorial nature of biofilm formation and the multiple potential targets of sRNA molecules, other approaches are required to more accurately determine the pathways and genes affected. Current studies are underway to more precisely map each sRNA by determining the transcriptional start sites and stop sites using 5′ and 3′ RACE, and to verify putative mRNA targets. The exact role of these sRNA in the regulation of biofilm formation in A. actinomycetemcomitans will be explored through further mutant analysis and transcriptional profiling.
Supplementary Material
Acknowledgments
We would like to thank Dr. Brian Tjaden for incorporating A. actinomycetemcomitans genome into TargetRNA website. We would like to thank Dr. Natalie King for providing the FURTA strains, and to Dr. Ashu Sharma, and Dr. Steven Gill for technical advice. This research was supported in part by USPHS Training Grant DE007034 from the National Institute of Dental and Craniofacial Research.
References
- Achenbach LA, Genova EG. Transcriptional regulation of a second flavodoxin gene from Klebsiella pneumoniae. Gene. 1997;194:235–240. doi: 10.1016/s0378-1119(97)00168-6. [DOI] [PubMed] [Google Scholar]
- Amarasinghe JJ, Scannapieco FA, Haase EM. Transcriptional and translational analysis of biofilm determinants of Aggregatibacter actinomycetemcomitans in response to environmental perturbation. Infect Immun. 2009;77:2896–2907. doi: 10.1128/IAI.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashova NV, Crosby JA, Al Ghofaily L, Kachlany SC. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect Immun. 2006a;74:2015–2021. doi: 10.1128/IAI.74.4.2015-2021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashova NV, Diaz R, Balashov SV, Crosby JA, Kachlany SC. Regulation of Aggregatibacter (Actinobacillus) actinomycetemcomitans leukotoxin secretion by iron. J Bacteriol. 2006b;188:8658–8661. doi: 10.1128/JB.01253-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender KS, Yen HC, Hemme CL, Yang Z, He Z, He Q, et al. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 2007;73:5389–5400. doi: 10.1128/AEM.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee MK, Fine DH, Figurski DH. tfoX (sxy)-dependent transformation of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene. 2007;399:53–64. doi: 10.1016/j.gene.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Garrido E, Llagostera M, Perez de Rozas AM, Badiola I, Barbe J. Pasteurella multocida exbB, exbD and tonB genes are physically linked but independently transcribed. FEMS Microbiol Lett. 2002;210:201–208. doi: 10.1111/j.1574-6968.2002.tb11181.x. [DOI] [PubMed] [Google Scholar]
- Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, et al. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Bio Systems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine DH, Velliyagounder K, Furgang D, Kaplan JB. The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and old world primates. Infect Immun. 2005;73:1947–1953. doi: 10.1128/IAI.73.4.1947-1953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KP, Chung WO, Lamont RJ, Demuth DR. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect Immun. 2001;69:7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KP, Gao L, Demuth DR. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect Immun. 2003;71:298–308. doi: 10.1128/IAI.71.1.298-308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, et al. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Straus NA. Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology. 1996;142 ( Pt 6):1469–1476. doi: 10.1099/13500872-142-6-1469. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Stealth regulation: biological circuits with small RNA switches. Genes Dev. 2002;16:2829–2842. doi: 10.1101/gad.1030302. [DOI] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol. 2004a;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Small RNAs shed some light. Cell. 2004b;118:1–2. doi: 10.1016/j.cell.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, et al. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc Natl Acad Sci U S A. 2003;100:9542–9547. doi: 10.1073/pnas.1033001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- Haase EM, Bonstein T, Palmer RJ, Jr, Scannapieco FA. Environmental influences on Actinobacillus actinomycetemcomitans biofilm formation. Arch Oral Biol. 2006;51:299–314. doi: 10.1016/j.archoralbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Haase EM, Stream JO, Scannapieco FA. Transcriptional analysis of the 5′ terminus of the flp fimbrial gene cluster from Actinobacillus actinomycetemcomitans. Microbiology. 2003;149:205–215. doi: 10.1099/mic.0.25786-0. [DOI] [PubMed] [Google Scholar]
- Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- Haraszthy VI, Lally ET, Haraszthy GG, Zambon JJ. Molecular cloning of the fur gene from Actinobacillus actinomycetemcomitans. Infect Immun. 2002;70:3170–3179. doi: 10.1128/IAI.70.6.3170-3179.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Tanimoto I, Ohta H, Kato K, Murayama Y, Fukui K. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol Immunol. 1998;42:253–258. doi: 10.1111/j.1348-0421.1998.tb02280.x. [DOI] [PubMed] [Google Scholar]
- Inoue T, Tanimoto I, Tada T, Ohashi T, Fukui K, Ohta H. Fermentable-sugar-level-dependent regulation of leukotoxin synthesis in a variably toxic strain of Actinobacillus actinomycetemcomitans. Microbiology. 2001;147:2749–2756. doi: 10.1099/00221287-147-10-2749. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, et al. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AH, Weber DJ, Oddone EZ, Perfect JR. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev Infect Dis. 1989;11:46–63. doi: 10.1093/clinids/11.1.46. [DOI] [PubMed] [Google Scholar]
- Kaplan JB, Meyenhofer MF, Fine DH. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J Bacteriol. 2003;185:1399–1404. doi: 10.1128/JB.185.4.1399-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AE, Metzger DJ, Murphy ER, Connell TD. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect Immun. 2001;69:6951–6961. doi: 10.1128/IAI.69.11.6951-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. A novel Fur- and iron-regulated small RNA, NrrF, is required for indirect Fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J Bacteriol. 2007;189:3686–3694. doi: 10.1128/JB.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun. 2005a;73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect Immun. 2005b;73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Procedures for working with lac. In: Miller JH, editor. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Press; 1992. [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najimi M, Lemos ML, Osorio CR. Identification of siderophore biosynthesis genes essential for growth of Aeromonas salmonicida under iron limitation conditions. Appl Environ Microbiol. 2008;74:2341–2348. doi: 10.1128/AEM.02728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio CR, Lemos ML, Braun V. Identification of Fur-regulated genes in the bacterial fish pathogen Photobacterium damselae ssp. piscicida using the Fur titration assay. Biometals. 2004;17:725–733. doi: 10.1007/s10534-004-1652-7. [DOI] [PubMed] [Google Scholar]
- Rhodes ER, Menke S, Shoemaker C, Tomaras AP, McGillivary G, Actis LA. Iron acquisition in the dental pathogen Actinobacillus actinomycetemcomitans: what does it use as a source and how does it get this essential metal? Biometals. 2007;20:365–377. doi: 10.1007/s10534-006-9058-3. [DOI] [PubMed] [Google Scholar]
- Rhodes ER, Tomaras AP, McGillivary G, Connerly PL, Actis LA. Genetic and functional analyses of the Actinobacillus actinomycetemcomitans AfeABCD siderophore-independent iron acquisition system. Infect Immun. 2005;73:3758–3763. doi: 10.1128/IAI.73.6.3758-3763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe BA, Najar FZ, Gillaspy A, Clifton S, Ducey T, Lewis L, et al. This project is supported by USPHS/NIH grant from the National Institute of Dental Research. Actinobacillus Genome Sequencing Project. [Google Scholar]
- Rosan B, Slots J, Lamont RJ, Listgarten MA, Nelson GM. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol Immunol. 1988;3:58–63. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- Scannapieco FA, Millar SJ, Reynolds HS, Zambon JJ, Levine MJ. Effect of anaerobiosis on the surface ultrastructure and surface proteins of Actinobacillus actinomycetemcomitans (Haemophilus actinomycetemcomitans) Infect Immun. 1987;55:2320–2323. doi: 10.1128/iai.55.9.2320-2323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia EA, Kuehner DM, Figurski DH. Mechanism of retrotransfer in conjugation: prior transfer of the conjugative plasmid is required. J Bacteriol. 1996;178:1457–1464. doi: 10.1128/jb.178.5.1457-1464.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan PK, LeBlanc DJ, Lee LN, Fives-Taylor P. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect Immun. 1991;59:4621–4627. doi: 10.1128/iai.59.12.4621-4627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Hantke K. Fur regulon in Gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson VJ, Bhattacharjee MK, Fine DH, Derbyshire KM, Figurski DH. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J Bacteriol. 1999;181:7298–7307. doi: 10.1128/jb.181.23.7298-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, et al. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Long T, Svenningsen SL, Wingreen NS, Bassler BL. Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol Cell. 2010;37:567–579. doi: 10.1016/j.molcel.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Gerhart E, Wagner H. Approaches to Identify Novel Non-messenger RNAs in Bacteria and to Investigate their Biologic Functions: RNA Mining. In: Hartmann RK, Binderreif A, Schon A, Westhof E, editors. Handbook of RNA Biochemistry. Weinheim: Wiley-VCH Verlag GmbH and Co; 2009. pp. 603–603. [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, et al. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen PT, Vulto I, Boxem M, de Graaff J. Characterization of a periplasmic protein involved in iron utilization of Actinobacillus actinomycetemcomitans. J Bacteriol. 1997;179:4949–4952. doi: 10.1128/jb.179.15.4949-4952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.