Abstract
Biohydrogen production (BHP) can be achieved by direct or indirect biophotolysis, photo-fermentation and dark fermentation, whereof only the latter does not require the input of light energy. Our motivation to compile this review was to quantify and comprehensively report strains and process performance of dark fermentative BHP. This review summarizes the work done on pure and defined co-culture dark fermentative BHP since the year 1901. Qualitative growth characteristics and quantitative normalized results of H2 production for more than 2000 conditions are presented in a normalized and therefore comparable format to the scientific community.
Statistically based evidence shows that thermophilic strains comprise high substrate conversion efficiency, but mesophilic strains achieve high volumetric productivity. Moreover, microbes of Thermoanaerobacterales (Family III) have to be preferred when aiming to achieve high substrate conversion efficiency in comparison to the families Clostridiaceae and Enterobacteriaceae.
The limited number of results available on dark fermentative BHP from fed-batch cultivations indicates the yet underestimated potential of this bioprocessing application. A Design of Experiments strategy should be preferred for efficient bioprocess development and optimization of BHP aiming at improving medium, cultivation conditions and revealing inhibitory effects. This will enable comparing and optimizing strains and processes independent of initial conditions and scale.
Keywords: Hydrogen yield, Volumetric hydrogen productivity, Specific hydrogen productivity, Closed batch, Batch, Chemostat culture, Fed-batch, Bioprocess, Result normalization, Scalable physiological parameters
Introduction
A possibility to circumvent the production of non-carbon neutral greenhouse gasses, such as carbon dioxide (CO2), is the development and continuous investigation of alternative biofuels. One promising alternative fuel is biologically generated hydrogen (H2), which is referred to as biohydrogen. Biohydrogen production (BHP) has initially been described in the 19th century [1,2], but elucidation of dark fermentative biohydrogen production was only done since 1901 [3,4]. Dark fermentative BHP is a carbon neutral process for production of H2 and CO2 from biomass by facultative and obligate anaerobic microorganisms.
H2 offers many beneficial features, such as being harmless to mammals and the environment [5-7]. Moreover, H2 is regarded as a non-polluting fuel, because its combustion product is water (H2O). When comparing volumetric energy densities at normal conditions for H2 and methane (CH4) a drawback of H2 is emerging, since H2 comprises 3.00 kWh m-3 in comparison to 9.97 kWh m-3 for CH4[8]. Moreover, H2 storage and transmission problems have been addressed [6,9-12], but research for optimization is ongoing [13].
H2 may be produced by a number of different processes including electrolysis of H2O, thermocatalytic reformation of hydrogen rich substrates and various biological processes [14-18]. To date, H2 is mainly produced by electrolysis of H2O and steam reformation of CH4. These processes are very energy intensive, but can be simply performed. Biological processes can be carried out at ambient temperatures, but they are more sophisticated in design and performance [18,19]. BHP can be achieved by direct or indirect biophotolysis, photo-fermentation and dark fermentation [15,16,18,20-25].
The main advantage of dark fermentative BHP is that the hydrogen evolution rate (HER) [mmol L-1 h-1 is higher in contrast to other BHP processes [11,12,17,26]. Major known drawbacks of dark fermentative BHP are the low yield of H2 per substrate consumed (Y(H2/S)) [mol mol-1, which is due to metabolic fundamentals [27]. Moreover, concomitant production of carbon rich metabolites (i.e. organic acids, alcohols) and CO2 is shown [28] and must be individually evaluated for each strain. CO2 can be removed or separated from H2, sequentially stored in biomass [29], or converted to other substances, such as CH4[30,31]. Basic microbiological investigations and bioprocess engineering research was performed to increase the overall strain performance of BHP during fermentation of pure microorganisms [32-34].
For a structured approach, we classified the vast amount of information available in literature in respect to the process modes, such as closed batch, batch, chemostat culture and fed-batch conditions and quantified the main performance attributes. We reviewed the exciting work published on microbial strains capable of dark fermentative BHP with the aim to demonstrate the versatile portfolio of H2 producing genera and the wide range of possible substrates for this purpose. In the present contribution the Y(H2/S), HER and specific hydrogen productivity (qH2) [mmol g-1 h-1] of the families Clostridiaceae, Enterobacteriaceae and Thermoanaerobacterales (Family III), as well as mesophilic and thermophilic cultivation conditions have been statistically compared. We analyse the microbiological and biochemical engineering approaches for optimization of H2 production and provide a comprehensive summary of the current status of dark fermentative BHP from more than 2000 different conditions. Herewith we want to stress that more quantitative work is urgently needed to turn this natural capacity into economic processes, based on physiological scalable parameters, which allow comparison and targeted optimization. Therefore, as a significant contribution to future work we propose a set of physiological scalable parameters for normalized results.
Classification and quantification tasks
In order to show a complete picture of each strain’s H2 production potential the results of dark fermentative BHP are presented in Additional files 1, 2, 3, 4, 5. Qualitative and quantitative characteristics are summarized as follows: taxonomic classification (genus, species, strain), quantitative performance attributes of BHP (Y(H2/S) [mol mol-1] (substrate conversion efficiency), the HER [mmol L-1 h-1] (volumetric productivity) and the qH2 [mmol g-1 h-1] (biological production capacity)), and qualitative attributes (i.e. pH, temperature and substrate). Moreover, we introduce a new categorization system in order to subclassify results according to the experimental set-up. This was required since many experiments have been conducted in sealed vials. We denote this cultivation technique as “closed batch”. This is a very prominent microbiological cultivation technique, which has to be distinguished from batch cultivation in open systems ( i.e. bioreactors). Thereafter, the following categorization was used: batch, chemostat culture and fed-batch.
Many authors stress for the importance to uniformly present yield and rates of BHP [11,12,35]. Result comparison is most suitably to be achieved by using culture dependent Y(H2/S) and qH2, as well as the non-culture dependent HER, because these units completely describe the strains H2 production characteristics. Moreover, the yield and rates are independent of scale and initial process conditions. We are certainly aware of the fact that presentation of results is even more advantageous based on a C-molar basis of the substrate [36]. Therefore, experiments have to be performed on defined media rather than on complex media, because the calculation of C-molar yield and productivities is not possible when analysing the performance in complex media. Thus, sophisticated analysis methods need to be considered for the evaluation of Y(H2/S), HER and qH2 based on C-molar mass balance. In this respect, we want to generally stress the importance of result presentation using mass balances, which is very important for quality assurance and must not be omitted [37]. Complete quantitative comparison of dark fermentative BHP would become possible if a C-molar basis of result presentation is used throughout the scientific community.
Review
Pure and defined co-culture experiments
A summarization the distribution of quantitative results obtained from different experimental set-ups of dark fermentative BHP is shown in Table 1. It becomes obvious that Y(H2/S) is most often presented, which is followed by the HER, whereas qH2 is described in less extent. Most studies on Y(H2/S) or HER were performed by either closed batch or batch fermentation (Additional files 12). A special case represents the fed-batch fermentation, whereof only five results for Y(H2/S) can be found in literature [38]. Results of BHP from defined co-culture examinations are also presented within the Additional files 1234.
Table 1.
Overview of dark fermentative BHP in respect to the cultivation technique
| Culture parameter | Closed batch | Batch | Chemostat culture | Fed-batch |
|---|---|---|---|---|
| Y(H2/S) |
441 |
425 |
253 |
5 |
| HER |
329 |
333 |
171 |
0 |
| qH2 | 68 | 78 | 99 | 0 |
Non-quantitative H2 production
Many dark fermentative BHP strains were isolated and characterized, but quantitative information on H2 production is missing. These strains and their corresponding growth requirements can serve as a pool to extend microbiological and bioprocess engineering examinations to new taxa. Furthermore, we are often confronted with the fact that quantitative results are assessed, but cannot be normalized by using the units Y(H2/S), HER or qH2, because of missing or undefined entities for recalculation of presented results. Consequently, these results and corresponding conditions are assigned to Additional file 1, because we have not been able to normalize these results for comparison purposes.
Closed batch H2 production
Dark fermentative BHP is found to be most often performed by strain cultivation in closed vessels (Additional file 2). Closed batch technique offers the main advantage that highly sophisticated bioprocess cultivation set-up for research can be omitted. Moreover, simple incubation conditions may be easily accomplished, because only incubation in H2O or air bath is necessary. In our opinion a closed batch investigation is highly advantageous in order to examine physical factors affecting BHP. For instance the elucidation of optimum temperature values, the effect of gas pressure, the influence of illumination or the investigation of agitation can be investigated. In this respect the inhibition of CO2 and H2 was described [39,40]. Additionally, by using closed batch technique, the substrate utilization spectrum can be investigated. This mode has the advantage to screen fast, determine optimal physical parameters, and describe their relationship to the physiological performance. Hence, the application closed batch is indeed of great value. The elucidation of chemical factors on BHP, such as the pH value seems to be rather difficult, because balanced growth at a certain pH value by means of base addition cannot be simply achieved. The investigation of the initial substrate concentration and medium amendments in order to optimize medium composition has been conducted, and the results led to an optimized medium composition [41,42].
A disadvantage of the closed batch technique is the discontinuous monitoring of culture parameters and the occurrence of unstable culture conditions due to sample removal and/or inhibition of BHP by build up of liquid and gaseous metabolic end products, because these excreted cellular end products cannot be continuously removed from the closed culture vessel. Manipulation to the culture vessel or to the culture itself requires at least the disruption of one physical factor. This unavoidably results in non-continuous cultivation conditions, making the utilization of closed batch technique rather unattractive, if sampling occurs more than once, because the culture response to changes in environmental conditions occurs rapidly [43]. Considering advantages and disadvantages of closed batch investigation the most urgent question to be addressed is: how quantitative is closed batch? Although, balanced growth may not be achieved, H2 production and growth kinetics were successfully investigated using closed batch technique [41,42]. End product inhibition occurring during closed batch investigation resulting from the production of solvents, organic acids, alcohols, CO2 or H2 partial pressure build-up certainly influences the results [39,40]. Hence, closed batch systems can be used for fast screening, but open cultivation systems need to be used for subsequent examination of the physiological potential of the strain and for quantitative bioprocess development.
Batch, chemostat culture and fed-batch H2 production
We provide an overview of dark fermentative BHP in bioreactors and similar set-ups, such as modified Erlenmeyer-flasks and refer to these examination techniques as open systems, because gas sparging, offgas composition determination, pH titration and medium supplementation can be performed. By using a highly controlled and automated set-up it is possible to quantitatively describe the strains inherent H2 production capacity and growth kinetics. Usually, this is performed by using fully automated and controlled bioreactor set-ups [34,44]. We compare the strains based on their BHP potential on glucose, and do not distinguish between growth on complex or defined medium. Furthermore, the pH value and temperature was not taken into account for comparison purposes.
Batch H2 production
Many quantitative investigations related to biohydrogen production were conducted by using batch type fermentations. Hereof, the genera BacillusCaldicellulosiruptorClostridiumEnterobacter and Escherichia were most widely studied (Additional file 3). Based on Y(H2/S) we identified Caldicellulosiruptor owensensis DSM 13100 [44] and Enterobacter cloacae DM 11 [33] showing highest Y(H2/S) of 4.0 and 3.9 mol mol-1, respectively. The highest HER of 32 mmol L-1 h-1 is shown for Enterobacter cloacae II BT-08 [32], which is followed by Clostridium sp. strain no. 2 showing a HER of 27 mmol L-1 h-1[45]. When analysing results for the highest qH2 we reveal that Caldicellulosiruptor saccharolyticus DSM 8903 produces 23 mmol g-1 h-1[28].
Chemostat culture H2 production
Dark fermentative BHP has often been investigated in chemostat culture. The results are summarized in Additional file 4. Thereof Caldicellulosiruptor saccharolyticus DSM 8903 is identified to comprise the highest Y(H2/S) of 4.0 mol mol-1[46]. Highest HER of 77 mmol L-1 h-1 is reported for Enterobacter cloacae II BT-08 [47]. The highest qH2 of 35 mmol g-1 h-1 is identified for Caldicellulosiruptor kristjanssonii DSM 12137 [34].
Fed-batch H2 production
The literature survey of dark fermentative BHP revealed only five conditions which have been operated in fed-batch mode (Additional file 5). The quantitative presentation of these results is restricted to the Y(H2/S). The maximum Y(H2/S) is identified for a recombinant strain of ATCC 25755 comprising 2.15 mol mol-1[38]. The limited number of results available for dark fermentative BHP from fed-batch fermentation is due to the fact that usually the application of this technique leads to massive accumulation of organic acids and other reduced end products (i.e. alcohols) in the culture broth, strongly inhibiting the growth and H2 production kinetics. Consequently, fed-batch investigation can be applied only by using broth exchange or cell separation systems [38]. Based on such experimental set-ups high cell densities and feed flow rates above the maximum specific growth rate can be reached [36]. The potential of fed-batch cultivation for H2 production is yet underestimated in terms of quantity and quality (Additional file 5). Thus, the quantitative potential of fed-batch cultivation has to be exploited in more detail for dark fermentative BHP by using biochemical engineering principles.
Discussion
Comparison of H2 production performance of strains related to Clostridiaceae, Enterobacteriaceae and Thermoanaerobacterales (Family III)
As summarized in Additional files 2345 quantitative examination of dark fermentative BHP is largely performed on strains phylogenetically related to either the family Clostridiaceae or Enterobacteriaceae, but also strains belonging to the family Thermoanaerobacterales (Family III) receive increasing scientific attention, because they comprise certain beneficial metabolic features [46,48-51]. According to Table 2 less results for qH2 than for HER compared to the Y(H2/S) are described. This discrepancy in the number of results available in literature is interesting, because during research the determination of biomass concentration and H2 offgas content could be easily performed. We analysed ClostridiaceaeEnterobacteriaceae and Thermoanaerobacterales (Family III) in order to elucidate differences of the performance of these families concerning HER and qH2 in respect to Y(H2/S). The basis for comparison is either any carbon substrate (Figure 1) or glucose (Figure 2), but irrespectively of growth conditions and metabolic modifications.
Table 2.
Results of the statistical analysis forClostridiaceae,EnterobacteriaceaeandThermoanaerobacterales(Family III)
|
Y(H2/S) |
Clostridiaceae (n = 464) |
Enterobacteriaceae (n = 295) |
Thermoanaerobacterales (Family III) (n = 73) |
| [mol mol-1] |
|
|
|
| Median |
1.785 |
0.82 |
2.9 |
| Mean |
1.87 ± 1.10* |
1.15 ± 1.34* |
2.92 ± 1.18* |
|
HER |
Clostridiaceae (n = 317) |
Enterobacteriaceae (n = 318) |
Thermoanaerobacterales (Family III) (n = 38) |
| [mmol L-1 h-1] |
|
|
|
| Median |
8.67 |
4.915 |
9.6 |
| Mean |
9.75 ± 8.41 |
11.37 ± 17.71 |
8.98 ± 3.40 |
|
qH2 |
Clostridiaceae (n = 70) |
Enterobacteriaceae (n = 102) |
Thermoanaerobacterales (Family III) (n = 20) |
| [mmol g-1 h-1] |
|
|
|
| Median |
10.05 |
3.75 |
16.615 |
| Mean | 14.49 ± 11.24 | 12.90 ± 26.02 | 18.61 ± 7.14 |
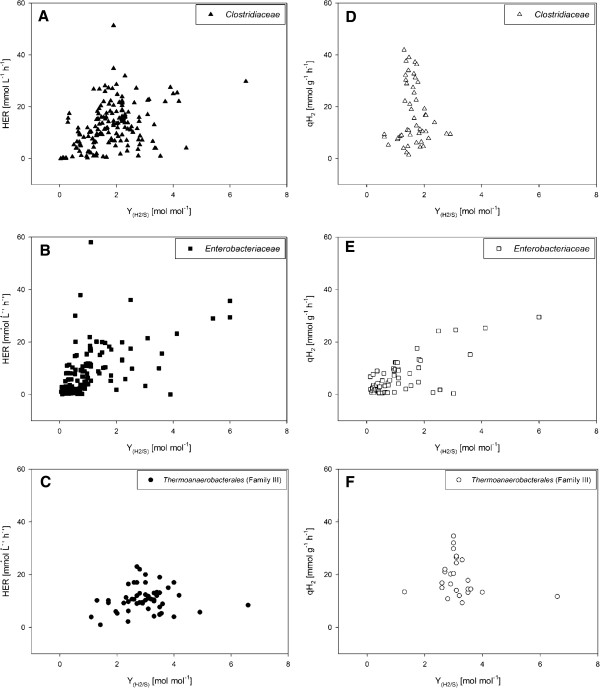
Figure 1 .
A graphical overview is shown irrespective of the utilization of the carbon substrate, medium composition and cultivation conditions of the HER ofClostridiaceae(A),Enterobacteriaceae(B) andThermoanaerobacterales(Family III) (C) plotted against the Y(H2/S). The qH2ofClostridiaceae(D),Enterobacteriaceae(E) andThermoanaerobacterales(Family III) (F) is shown in relation to the Y(H2/S). It is indicated that Clostridiaceae and Enterobacteriaceae perform better than Thermoanaerobacterales (Family III) in respect to the volumetric productivity. The qH2 for Clostridiaceae and Thermoanaerobacterales (Family III) is shown to be higher than for Enterobacteriaceae. Regarding the substrate conversion efficiency (Y(H2/S)) the following ranking is indicated: Thermoanaerobacterales (Family III) > Clostridiaceae > Enterobacteriaceae.
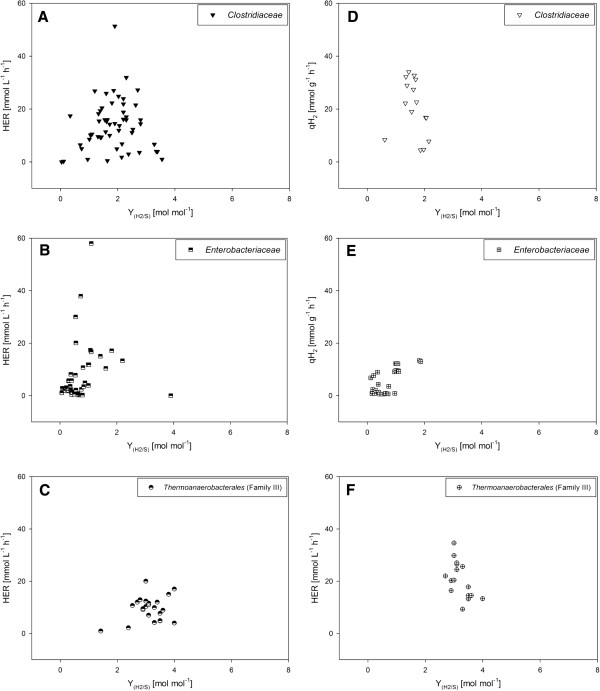
Figure 2 .
A graphical overview of the utilization of glucose is presented. HER ofClostridiaceae(A),Enterobacteriaceae(B) andThermoanaerobacterales(Family III) (C) is shown in relation to the Y(H2/S). The qH2ofClostridiaceae(D),Enterobacteriaceae(E) andThermoanaerobacterales(Family III) (F) in relation to the Y(H2/S)is also presented. These results are depicted irrespective of the medium composition, cultivation conditions or genetic modification. These graphs offer an indication that Clostridiaceae and Enterobacteriaceae show a higher volumetric productivity than Thermoanaerobacterales (Family III). In respect to the qH2Clostridiaceae and Thermoanaerobacterales (Family III) show higher productivity than Enterobacteriaceae. Based on the Y(H2/S) following ranking is presented: Thermoanaerobacterales (Family III) > Clostridiaceae > Enterobacteriaceae.
These three culture parameters are plotted against each other on any carbon substrate (Figures 1A-1F). Based on the HER Clostridiaceae and Enterobacteriaceae comprise highest volumetric productivity, whereas in respect to the specific H2 productivity Clostridiaceae and Thermoanaerobacterales (Family III) are indicated to perform better. Nonetheless, a clear trend towards better substrate conversion efficiency is revealed: Thermoanaerobacterales (Family III) > Clostridiaceae > Enterobacteriaceae. Secondly, the HER, qH2 and Y(H2/S) are graphically analysed for the growth of the three families on glucose, but independent of the utilization of complex or defined medium, cultivation conditions or metabolic modifications (Figures 2A-2F). In principle analogous trends for the HER, qH2 and Y(H2/S) of the three families can be shown for the growth on glucose compared to the growth on any carbon substrate.
Above paragraph described the interrelations between the physiological parameters of ClostridiaceaeEnterobacteriaceae and Thermoanaerobacterales (Family III). Subsequently, we want to statistically evaluate their dark fermentative BHP potential. Firstly, a comparison has been done based on the median in order to identify a superior performance of one of the three families regarding the Y(H2/S), HER and qH2[52]. Secondly, the mean between these families has been individually analysed by using the Welch-test, or where applicable by using the Student’s t-test, at a level of significance of p = 0.01 [52]. Normalized results available in the Additional files 2345 have been used for comparison purposes. The results are summarized in Table 2.
By comparing the median in respect to the Y(H2/S), HER and qH2 we can show that strains perform as follows: Thermoanaerobacterales (Family III) > Clostridiaceae > Enterobacteriaceae. Statistical analysis of the mean only allows a most significant statement for the Y(H2/S), but not for the HER and qH2. Herewith we present evidence that strains of Thermoanaerobacterales (Family III) perform most significantly better in respect to the Y(H2/S) of Clostridiaceae and Enterobacteriaceae. Also strains from the family Clostridiaceae are found to comprise a most significantly higher Y(H2/S) than strains of the family Enterobacteriaceae. Based on our statistical investigation we reveal that strains of the family Thermoanaerobacterales (Family III) have to be clearly preferred when aiming to achieve a high Y(H2/S) in comparison to the families Clostridiaceae and Enterobacteriaceae.
Comparison of mesophilic and thermophilic H2 production
In order to compare the dark fermentative BHP performance of mesophilic (20-44 °C) and thermophilic (45-80 °C) strains we used the results shown in Additional files 2, 3, 4, 5. As mentioned above statistical analysis has been carried out to evaluate the difference in terms of Y(H2/S), qH2 and HER. The results are summarized in Table 3. We can clearly show that thermophilic strains are superior to mesophilic strains in respect to the Y(H2/S). This result is also supported in the values for the median, which also shows the higher Y(H2/S) of thermophilic strains. Herewith most significant evidence is presented to favour mesophilic over thermophilic strains in respect to the HER, which is also reflected in the value determined for the median. Unfortunately our statistical analysis of qH2 cannot present a beneficial result of one or the other group, but nevertheless the mean and the median show a trend to favour thermophilic over mesophilic strains. Thus, we are able to present statistical evidence demonstrating to use thermophilic strains when aiming on a high Y(H2/S), but to use mesophilic strains to achieve a high HER.
Table 3.
Statistical analysis of mesophilic and thermophilc dark fermentative BHP
|
Y(H2/S) |
Mesophilic |
Thermophilic |
| [mol mol-1] |
(n = 695) |
(n = 244) |
| Median |
1.22 |
2.30 |
| Mean |
1.46 ± 1.18* |
2.20 ± 1.42* |
|
HER |
Mesophilic |
Thermophilic |
| [mmol L-1 h-1] |
(n = 587) |
(n = 128) |
| Median |
6.29 |
3.89 |
| Mean |
9.92 ± 13.27* |
5.62 ± 5.45* |
|
qH2 |
Mesophilic |
Thermophilic |
| [mmol g-1 h-1] |
(n = 147) |
(n = 50) |
| Median |
7.65 |
9.70 |
| Mean | 11.53 ± 16.00 | 12.51 ± 10.79 |
Microbiological potential for enhancing H2 production
Microbial potential of H2 production by strain isolation
The initial microbiological investigation of strains for biological H2 production offers the opportunity to characterize the microbes in full detail, for instance in respect to pH, temperature and substrate utilization spectrum. Moreover, phylogenetical information will eventually be retrieved during strain characterization. We suggest using the information on strains and conditions available in Additional files 12345 to extend studies of dark fermentative BHP to broader substrate diversity. These strains offer promising experimental endeavours to microbiologists for physiological studies, because many of these strains are yet not characterized in detail [53-56]. Many wild-type strains were found to comprise a high Y(H2/S) and high qH2[44,51,57]. Moreover, basic research efforts need to be increased to isolate novel dark fermentative BHP strains from the environment, because up to date only few of the estimated existing microbes have yet become cultivable [58,59]. Still the optimization of dark fermentative BHP by using wild-type microbes is an alternative.
Evaluating the microbial H2production potential by application of in silico analysis
Another highly noteworthy field related to dark fermentative BHP is the strain identification based on in silico analysis. The information gain is not restricted to phylogenetical knowledge, but also sequence information on enzymes is available [60,61]. Therefore, substantial information on the catalytic units for H2 production can be retrieved. Moreover, screening for specific enzymes in respect to substrate breakdown and utilization can also be done. Isolated information on certain microorganisms can be retrieved, but also whole genomes of several dark fermentative BHP strains are sequenced and provide full access for physiological and in silico analysis, offering putative modification possibilities towards metabolic engineering objectives.
Optimization of H2 production by application of metabolic engineering
Metabolic engineering is especially important for dark fermentative BHP strains that comprise high Y(H2/S) and qH2, but whereof high volumetric production rates are either inherently limited by metabolic bottlenecks (i.e. organic acids, solvent and alcohol production) or, when concerning thermophilic strains, by their achievable cell densities. Usually Escherichia coli is the target for metabolic engineering [62-68]. Since Clostridia spp. show high Y(H2/S) and high qH2 in comparison to Escherichia spp., metabolic strain engineering is an interesting option in order to increase HER. Nevertheless, Escherichia spp. can be genetically modified relatively easy in respect to their facultative anaerobic growth characteristic [64,65,67,69], hence, allowing unsophisticated achievable growth in a variety of culture vessels. Clostridia spp., Caldicellulosiruptor spp. and other strict anaerobic genera in turn require more sophisticated cultivation set-up, because they are obligate anaerobes. Nevertheless, strains of both genera have been genetically modified [64,65,67,69-72]. In this respect the application of directed evolution [73] towards optimization of cultivation conditions or the substrate utilization spectrum could be another favourable approach.
Biochemical engineering potential for increasing H2 production
Optimization of H2 production by bioprocess engineering
For a robust and commercial usefully application of dark fermentative BHP several factors have to be addressed. Firstly, chemical (i.e. pH, ionic strength, CO2 solubility) and physical factors (i.e. temperature, partial pressure of H2 and CO2, agitation) influencing the Y(H2/S) and qH2 need to be identified, which are usually already known and differ between various strains [28,40,74-77]. By using open cultivation systems removal of inhibitory gaseous compounds could be and is done by continuous stripping with inert gas. Secondly, factors for increasing the HER (cell retention, end product inhibition) have to be elucidated. In order to enhance the HER, an increase of the biomass concentration is required. This may be accomplished by using membrane filtration to separate unwanted metabolites and retain the biomass within the bioreactor. Hence, fed-batch cultivation for dark fermentative BHP can become a promissing approach.
The medium contains the carbon substrate for biomass and H2 production and is a very important starting point for optimization during bioprocess development. Many conditions, which are presented in Additional files 1, 2, 3, 4, 5, do not properly reflect the status of a pure carbon source for H2 production. Hence, in many experiments complex medium amendments are used. Since most of the undefined compounds undergo temporal fluctuations from lot to lot during the production process, its composition is not always consistent. Hence, the use of complex compounds does not easily allow conclusions on the influence of the carbon source on H2 production. In order to establish a robust bioprocess quantitative work on defined medium needs to be performed for strain characterization. This is an important consideration in order to elucidate the strains growth parameters and inherent potential of H2 production.
Use of Design of Experiments strategy for optimization of H2 production
Many articles have analysed the impact of the medium composition in order to increase Y(H2/S), HER or qH2[35,78-82]. These investigations have been performed invariantly, thus by changing only one culture variable, but more and more examinations use the advantage of Design of Experiments (DoE), which has proven to be very successful [41,55,83-89]. This experimental strategy allows multivariate analysis by modification of several variables at one time, and moreover to optimize for the response(s) of interest. During the successive steps of a DoE application, optimization of BHP can be achieved by elucidation of medium components, but also on other products than H2, such as CO2, organic acids, solvents and alcohols or even other inhibitory compounds. Moreover, the influence of chemical and physical parameters on H2 production may be included in the investigation. Hence, a comprehensive DoE is much faster in identification of the optimal operation point, to be individually optimized for the bioprocess of interest. DoE screening and successive optimization results in an amended medium composition, identifies the corresponding culture parameters and concomitantly the optimum cultivation conditions for improved H2 production.
Our review shows the inherent potential and the need for quantitative investigation of pure culture dark fermentative BHP. Especially the elucidation of non-food substrates for H2 production is possibly of higher potential commercial applicability. From this point of view, the use of complex media for H2 production could rather represent a putative real case scenario. However, strain characterization is crucial and has to be performed in defined media for elucidation of the strain's full physiological potential. Herewith we propose a set of physiological scalable parameters for characterization and optimization of dark fermentative BHP strains by using bioprocessing. The first step should be a sound investigation by appication of DoE for elucidation of the following culture parameters: Y(H2/S), MER and qH2. In a successive investigation the addition of complex or undefined medium componets should to be investigated and compared in respect to initial elucidated culture parameters. Hence, future investigations in this field of bioprocessing could be rapidly completed.
Conclusions
This review summarizes the work done on pure and defined co-culture dark fermentative BHP since the year 1901. Qualitative growth characteristics and quantitative normalized results of H2 production for more than 2000 conditions are presented. Now these normalized and comparable results become available to the scientific community.
Statistically based evidence shows that thermophilic strains comprise high substrate conversion efficiency, but mesophilic strains achieve high volumetric productivity.
Microbes of Thermoanaerobacterales (Family III) have to be preferred when aiming to achieve a high Y(H2/S) in comparison to the families Clostridiaceae and Enterobacteriaceae, based on a comprehensive statistical substantiation.
The limited number of results available on dark fermentative BHP from fed-batch cultivations indicates the yet underestimated potential of this bioprocessing application.
For an efficient bioprocess development the optimization of H2 production by using DoE strategy for medium modification, cultivation condition improvement and inhibitory compound analysis should be preferred and a set of physiological scalable parameters is suggested.
Comparability of key culture parameters of dark fermentative BHP is of utmost importance and thus the following entities should be used for the presentation of results: Y(H2/S) [mol C-mol-1], HER [mmol L-1 h-1] and qH2 [mmol g-1 h-1].
Abbreviations
BHP, Biological hydrogen production; CH4, Methane; C-mol, Moles of carbon; CO2, Carbon dioxide; H2, Molecular hydrogen; H2O, Water; HER, Volumetric hydrogen production rate; qH2, Specific hydrogen production rate; Y(H2/S), Moles of hydrogen produced per moles of substrate consumed.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SR reviewed the literature on dark fermentative biohydrogen production, prepared the tables, figures, additional files, performed the statistical analysis, drafted the manuscript and coordinated the review. SR and CH contributed to the conception and design of the manuscript. CH helped to draft the manuscript. All authors have read and approve the final version of the manuscript.
Supplementary Material
Strains reported to produce biohydrogen without the possibility to calculate or retrieve quantitative results[3,4,6,7,48,80,82,90-204].
Closed batch dark fermentative biohydrogen production[4,29,35,39,42,53-55,64-67,69,78,82,83,85,86,98,120,130,131,137,142,166,180,182-184,192,194,198,205-314].
Batch dark fermentative biohydrogen production[28,32,33,45,49,56,57,62,63,65,66,72,76,78,79,81,84,90,96,110,115,315-369].
Chemostat culture dark fermentative biohydrogen production[29,34,46,47,56,75,77,96,118,122,124,125,228,257-260,279,286,291,292,331,353,356,370-400].
Fed-batch dark fermentative biohydrogen production[38].
Contributor Information
Simon Rittmann, Email: simon.rittmann@tuwien.ac.at.
Christoph Herwig, Email: christoph.herwig@tuwien.ac.at.
Acknowledgements
The authors greatly acknowledge Prof. Dr. Helga Stan-Lotter and Prof. Dr. Peter Holubar for helpful discussions. We want to thank Dr. Christian Dietzsch, DI Arne Seifert and Dr. Oliver Spadiut for critical comments on the manuscript.
References
- Hoppe-Seyler F. Ueber die Processe der Gährungen und ihre Beziehung zum Leben der Organismen: Erste Abhandlung. Arch Gesamte Physiol. 1875;12:1–17. [Google Scholar]
- Popoff L. Ueber die Sumpfgasgährung. Arch Gesamte Physiol. 1875;10:113–146. doi: 10.1007/BF01639928. [DOI] [Google Scholar]
- Harden A. The chemical action of Bacillus coli communis and similar organisms on carbohydrates and allied compounds. J Chem Soc Trans. 1901;79:610–628. [Google Scholar]
- Pakes WCC, Jollyman WH. The bacterial decomposition of formic acid into carbon dioxide and hydrogen. J Chem Soc Trans. 1901;79:386–391. [Google Scholar]
- Peraldo Bicelli L. Hydrogen: a clean energy source. Int J Hydrogen Energ. 1986;11:555–562. doi: 10.1016/0360-3199(86)90121-7. [DOI] [Google Scholar]
- Bockris JOM. The origin of ideas on a hydrogen economy and its solution to the decay of the environment. Int J Hydrogen Energ. 2002;27:731–740. doi: 10.1016/S0360-3199(01)00154-9. [DOI] [Google Scholar]
- Zajic JE, Margaritis A, Brosseau JD. Microbial hydrogen production from replenishable resources. Int J Hydrogen Energ. 1979;4:385–402. doi: 10.1016/0360-3199(79)90101-0. [DOI] [Google Scholar]
- Wasserstoff Daten - Hydrogen Data. http://www.h2data.de/
- Cherry RS. A hydrogen utopia? Int J Hydrogen Energ. 2003;29:125–129. [Google Scholar]
- Das D, Khanna N, Veziroglu TN. Recent developments in biological hydrogen production processes. Chem Ind Chem Eng Q. 2008;14:57–67. doi: 10.2298/CICEQ0802057D. [DOI] [Google Scholar]
- Levin DB, Pitt L, Love M. Biohydrogen production: prospects and limitations to practical application. Int J Hydrogen Energ. 2003;29:173–185. [Google Scholar]
- Levin DB, Pitt L, Love M. Biohydrogen production: prospects and limitations to practical application. [Erratum to document cited in CA140:166604] Int J Hydrogen Energ. 2004;29:1425–1426. doi: 10.1016/j.ijhydene.2004.05.004. [DOI] [Google Scholar]
- Boddien A, Mellmann D, Gaertner F, Jackstell R, Junge H, Dyson PJ, Laurenczy G, Ludwig R, Beller M. Efficient Dehydrogenation of Formic Acid Using an Iron Catalyst. Science. 2011;333:1733–1736. doi: 10.1126/science.1206613. [DOI] [PubMed] [Google Scholar]
- Momirlan M, Veziroglu T. Recent directions of world hydrogen production. Renew Sust Energ Rev. 1999;3:219–231. doi: 10.1016/S1364-0321(98)00017-3. [DOI] [Google Scholar]
- Lopes Pinto FA, Troshina O, Lindblad P. A brief look at three decades of research on cyanobacterial hydrogen evolution. Int J Hydrogen Energ. 2002;27:1209–1215. doi: 10.1016/S0360-3199(02)00089-7. [DOI] [Google Scholar]
- Nandi R, Sengupta S. Microbial production of hydrogen: an overview. Crit Rev Microbiol. 1998;24:61–84. doi: 10.1080/10408419891294181. [DOI] [PubMed] [Google Scholar]
- Hallenbeck PC, Benemann JR. Biological hydrogen production; fundamentals and limiting processes. Int J Hydrogen Energ. 2002;27:1185–1193. doi: 10.1016/S0360-3199(02)00131-3. [DOI] [Google Scholar]
- Das D, Veziroglu TN. Hydrogen production by biological processes: a survey of literature. Int J Hydrogen Energ. 2000;26:13–28. [Google Scholar]
- Nath K, Das D. Improvement of fermentative hydrogen production: various approaches. Appl Microbiol Biot. 2004;65:520–529. doi: 10.1007/s00253-004-1644-0. [DOI] [PubMed] [Google Scholar]
- Schuetz K, Happe T, Troshina O, Lindblad P, Leitao E, Oliveira P, Tamagnini P. Cyanobacterial H2 production - a comparative analysis. Planta. 2004;218:350–359. doi: 10.1007/s00425-003-1113-5. [DOI] [PubMed] [Google Scholar]
- Melis A. Green alga hydrogen production: progress, challenges and prospects. Int J Hydrogen Energ. 2002;27:1217–1228. doi: 10.1016/S0360-3199(02)00110-6. [DOI] [Google Scholar]
- Madamwar D, Garg N, Shah V. Cyanobacterial hydrogen production. World J Microb Biot. 2001;16:757–767. [Google Scholar]
- Benemann JR. Hydrogen production by microalgae. J Appl Phycol. 2000;12:291–300. doi: 10.1023/A:1008175112704. [DOI] [Google Scholar]
- Melis A, Melnicki MR. Integrated biological hydrogen production. Int J Hydrogen Energ. 2006;31:1563–1573. doi: 10.1016/j.ijhydene.2006.06.038. [DOI] [Google Scholar]
- Lee D-J, Show K-Y, Su A. Dark fermentation on biohydrogen production: Pure culture. Bioresource Technol. 2011;102:8393–8402. doi: 10.1016/j.biortech.2011.03.041. [DOI] [PubMed] [Google Scholar]
- Hallenbeck PC. Fundamentals of the fermentative production of hydrogen. Water Sci Technol. 2005;52:21–29. [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willquist K, Claassen PAM, van Niel EWJ. Evaluation of the influence of CO2 on hydrogen production by Caldicellulosiruptor saccharolyticus. Int J Hydrogen Energ. 2009;34:4718–4726. doi: 10.1016/j.ijhydene.2009.03.056. [DOI] [Google Scholar]
- Lo Y-C, Chen C-Y, Lee C-M, Chang J-S. Sequential dark-photo fermentation and autotrophic microalgal growth for high-yield and CO2-free biohydrogen production. Int J Hydrogen Energ. 2010;35:10944–10953. doi: 10.1016/j.ijhydene.2010.07.090. [DOI] [Google Scholar]
- Rittmann S, Seifert A, Herwig C. Quantitative analysis of media dilution rate effects on Methanothermobacter marburgensis grown in continuous culture on H2 and CO2. Biomass Bioenerg. 2012;36:293–301. [Google Scholar]
- Levin DB, Zhu H, Beland M, Cicek N, Holbein BE. Potential for hydrogen and methane production from biomass residues in Canada. Bioresource Technol. 2006;98:654–660. doi: 10.1016/j.biortech.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Khanna N, Kotay SM, Gilbert JJ, Das D. Improvement of biohydrogen production by Enterobacter cloacae IIT-BT 08 under regulated pH. J Biotechnol. 2011;152:9–15. doi: 10.1016/j.jbiotec.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Mandal B, Nath K, Das D. Improvement of Biohydrogen Production Under Decreased Partial Pressure of H2 by Enterobacter cloacae. Biotechnol Lett. 2006;28:831–835. doi: 10.1007/s10529-006-9008-8. [DOI] [PubMed] [Google Scholar]
- Zeidan AA, Raadstroem P, van Niel EWJ. Stable coexistence of two Caldicellulosiruptor species in a de novo constructed hydrogen-producing co-culture. Microb Cell Fact. 2010;9:102. doi: 10.1186/1475-2859-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova G, Rakhely G, Kovacs KL. Thermophilic biohydrogen production from energy plants by Caldicellulosiruptor saccharolyticus and comparison with related studies. Int J Hydrogen Energ. 2009;34:3659–3670. doi: 10.1016/j.ijhydene.2009.02.082. [DOI] [Google Scholar]
- Nielsen J, Villadsen J, Liden G. Bioreaction Engineering Principles. 2 2002. [Google Scholar]
- Herwig C, Marison I, Von Stockar U. On-line stoichiometry and identification of metabolic state under dynamic process conditions. Biotechnol Bioeng. 2001;75:345–354. doi: 10.1002/bit.10058. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhu Y, Yang S-T. Construction and Characterization of ack Deleted Mutant of Clostridium tyrobutyricum for Enhanced Butyric Acid and Hydrogen Production. Biotechnol Prog. 2006;22:1265–1275. doi: 10.1021/bp060082g. [DOI] [PubMed] [Google Scholar]
- Malik B, Su WW, Wald HL, Blumentals II, Kelly RM. Growth and gas production for the hyperthermophilic archaebacterium, Pyrococcus furiosus. Biotechnol Bioeng. 1989;34:1050–1057. doi: 10.1002/bit.260340805. [DOI] [PubMed] [Google Scholar]
- Park W, Hyun SH, Oh S-E, Logan BE, Kim IS. Removal of Headspace CO2 Increases Biological Hydrogen Production. Environ Sci Technol. 2005;39:4416–4420. doi: 10.1021/es048569d. [DOI] [PubMed] [Google Scholar]
- Jo JH, Lee DS, Park D, Park JM. Statistical optimization of key process variables for enhanced hydrogen production by newly isolated Clostridium tyrobutyricum JM1. Int J Hydrogen Energ. 2008;33:5176–5183. doi: 10.1016/j.ijhydene.2008.05.012. [DOI] [Google Scholar]
- Xu L, Ren N, Wang X, Jia Y. Biohydrogen production by Ethanoligenens harbinense B49: Nutrient optimization. Int J Hydrogen Energ. 2008;33:6962–6967. doi: 10.1016/j.ijhydene.2008.09.005. [DOI] [Google Scholar]
- Blackwell JR, Gilmour DJ. Physiological response of the unicellular green alga Chlorococcum submarinum to rapid changes in salinity. Arch Microbiol. 1991;157:86–91. [Google Scholar]
- Zeidan AA, van Niel EWJ. A quantitative analysis of hydrogen production efficiency of the extreme thermophile Caldicellulosiruptor owensensis OL. Int J Hydrogen Energ. 2010;35:1128–1137. doi: 10.1016/j.ijhydene.2009.11.082. [DOI] [Google Scholar]
- Taguchi F, Mizukami N, Hasegawa K, Saito-Taki T. Microbial conversion of arabinose and xylose to hydrogen by a newly isolated Clostridium sp. No. 2. Can J Microbiol. 1994;40:228–233. doi: 10.1139/m94-037. [DOI] [Google Scholar]
- de Vrije T, Mars AE, Budde MAW, Lai MH, Dijkema C, Waard P, Claassen PAM. Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biot. 2007;74:1358–1367. doi: 10.1007/s00253-006-0783-x. [DOI] [PubMed] [Google Scholar]
- Kumar N, Das D. Continuous hydrogen production by immobilized Enterobacter cloacae IIT-BT 08 using lignocellulosic materials as solid matrices. Enzyme Microb Technol. 2001;29:280–287. doi: 10.1016/S0141-0229(01)00394-5. [DOI] [Google Scholar]
- Herbel Z, Rakhely G, Bagi Z, Ivanova G, Acs N, Kovacs E, Kovacs KL. Exploitation of the extremely thermophilic Caldicellulosiruptor saccharolyticus in hydrogen and biogas production from biomasses. Environ Technol. 2009;31:1017–1024. doi: 10.1080/09593330.2010.484075. [DOI] [PubMed] [Google Scholar]
- Zeidan AA, Van Niel EWJ. Developing a thermophilic hydrogen-producing co-culture for efficient utilization of mixed sugars. Int J Hydrogen Energ. 2009;34:4524–4528. doi: 10.1016/j.ijhydene.2008.07.092. [DOI] [Google Scholar]
- van de Werken HJG, Verhaart MRA, VanFossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, van Niel EWJ. et al. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microb. 2008;74:6720–6729. doi: 10.1128/AEM.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willquist K, Zeidan AA, van Niel EWJ. Physiological characteristics of the extreme thermophile Caldicellulosiruptor saccharolyticus: an efficient hydrogen cell factory. Microb Cell Fact. 2010;9:89. doi: 10.1186/1475-2859-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datalab. http://www.lohninger.com/datalab/en_home.html.
- Ho K-L, Lee D-J. Harvesting biohydrogen from cellobiose from sulfide or nitrite-containing wastewaters using Clostridium sp. R1. Bioresource Technol. 2011;102:8547–8549. doi: 10.1016/j.biortech.2011.04.031. [DOI] [PubMed] [Google Scholar]
- Lo Y-C, Huang C-Y, Cheng C-L, Lin C-Y, Chang J-S. Characterization of cellulolytic enzymes and bioH2 production from anaerobic thermophilic Clostridium sp. TCW1. Bioresour Technol. 2011;102:8384–8392. doi: 10.1016/j.biortech.2011.03.064. [DOI] [PubMed] [Google Scholar]
- Pan CM, Fan YT, Xing Y, Hou HW, Zhang ML. Statistical optimization of process parameters on biohydrogen production from glucose by Clostridium sp. Fanp2. Bioresource Technol. 2008;99:3146–3154. doi: 10.1016/j.biortech.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Yokoi H, Aratake T, Hirose J, Hayashi S, Takasaki Y. Simultaneous production of hydrogen and bioflocculant by Enterobacter sp. BY-29. World J Microb Biot. 2001;17:609–613. doi: 10.1023/A:1012463508364. [DOI] [Google Scholar]
- van Niel EWJ, Budde MAW, de Haas GG, van der Wal FJ, Claassen PAM, Stams AJM. Distinctive properties of high hydrogen producing extreme thermophiles, Caldicellulosiruptor saccharolyticus and Thermotoga elfii. Int J Hydrogen Energ. 2002;27:1391–1398. doi: 10.1016/S0360-3199(02)00115-5. [DOI] [Google Scholar]
- Barer MR, Harwood CR. Bacterial viability and culturability. Adv Microb Physiol. 1999;41:93–137. doi: 10.1016/s0065-2911(08)60166-6. [DOI] [PubMed] [Google Scholar]
- Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol Ecol. 2006;15:1713–1731. doi: 10.1111/j.1365-294X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- Kalia VC, Lal S, Ghai R, Mandal M, Chauhan A. Mining genomic databases to identify novel hydrogen producers. Trends Biotechnol. 2003;21:152–156. doi: 10.1016/S0167-7799(03)00028-3. [DOI] [PubMed] [Google Scholar]
- Kalia VC, Purohit HJ. Microbial diversity and genomics in aid of bioenergy. J Ind Microbiol Biotechnol. 2008;35:403–419. doi: 10.1007/s10295-007-0300-y. [DOI] [PubMed] [Google Scholar]
- Redwood MD, Mikheenko IP, Sargent F, Macaskie LE. Dissecting the roles of Escherichia coli hydrogenases in biohydrogen production. FEMS Microbiol Lett. 2008;278:48–55. doi: 10.1111/j.1574-6968.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. Enhanced hydrogen production from formic acid by formate hydrogen lyase-overexpressing Escherichia coli strains. Appl Environ Microb. 2005;71:6762–6768. doi: 10.1128/AEM.71.11.6762-6768.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Sanchez-Torres V, Wood TK. Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl Microbiol Biot. 2007;77:879–890. doi: 10.1007/s00253-007-1217-0. [DOI] [PubMed] [Google Scholar]
- Maeda T, Sanchez-Torres V, Wood TK. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol. 2008;1:30–39. doi: 10.1111/j.1751-7915.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Sanchez-Torres V, Wood TK. Protein engineering of hydrogenase 3 to enhance hydrogen production. Appl Microbiol Biot. 2008;79:77–86. doi: 10.1007/s00253-008-1416-3. [DOI] [PubMed] [Google Scholar]
- Maeda T, Vardar G, Self WT, Wood TK. Inhibition of hydrogen uptake in Escherichia coli by expressing the hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803. BMC Biotechnol. 2007;7:25. doi: 10.1186/1472-6750-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardar-Schara G, Maeda T, Wood TK. Metabolically engineered bacteria for producing hydrogen via fermentation. Microb Biotechnol. 2008;1:107–125. doi: 10.1111/j.1751-7915.2007.00009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Torres V, Maeda T, Wood TK. Protein engineering of the transcriptional activator FhlA to enhance hydrogen production in Escherichia coli. Appl Environ Microb. 2009;75:5639–5646. doi: 10.1128/AEM.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pajuelo M, Meynial-Salles I, Mendes F, Soucaille P, Vasconcelos I. Microbial conversion of glycerol to 1,3-propanediol: Physiological comparison of a natural producer, Clostridium butyricum VPI 3266, and an engineered strain, Clostridium acetobutylicum DG1(pSPD5) Appl Environ Microb. 2006;72:96–101. doi: 10.1128/AEM.72.1.96-101.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Desvaux M, Petitdemange H. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl Environ Microb. 2002;68:53–58. doi: 10.1128/AEM.68.1.53-58.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, Kimura T, Sakka K, Ohmiya K. Overexpression of a hydrogenase gene in Clostridium paraputrificum to enhance hydrogen gas production. FEMS Microbiol Lett. 2005;246:229–234. doi: 10.1016/j.femsle.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Lin Z, Thorsen T, Arnold FH. Functional expression of horseradish peroxidase in E. coli by directed evolution. Biotechnol Prog. 1999;15:467–471. doi: 10.1021/bp990037r. [DOI] [PubMed] [Google Scholar]
- Wang J, Wan W. Factors influencing fermentative hydrogen production: A review. Int J Hydrogen Energ. 2009;34:799–811. doi: 10.1016/j.ijhydene.2008.11.015. [DOI] [Google Scholar]
- van Groenestijn JW, Geelhoed JS, Goorissen HP, Meesters KPM, Stams AJM, Claassen PAM. Performance and population analysis of a non-sterile trickle bed reactor inoculated with Caldicellulosiruptor saccharolyticus, a thermophilic hydrogen producer. Biotechnol Bioeng. 2009;102:1361–1367. doi: 10.1002/bit.22185. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Ren NQ, Xiang WS, Guo WQ. Influence of gaseous end-products inhibition and nutrient limitations on the growth and hydrogen production by hydrogen-producing fermentative bacterial B49. Int J Hydrogen Energ. 2007;32:748–754. doi: 10.1016/j.ijhydene.2006.08.003. [DOI] [Google Scholar]
- Collet C, Gaudard O, Peringer P, Schwitzguebel J-P. Acetate production from lactose by Clostridium thermolacticum and hydrogen-scavenging microorganisms in continuous culture-Effect of hydrogen partial pressure. J Biotechnol. 2005;118:328–338. doi: 10.1016/j.jbiotec.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Balint B, Bagi Z, Toth A, Rakhely G, Perei K, Kovacs KL. Utilization of keratin-containing biowaste to produce biohydrogen. Appl Microbiol Biot. 2005;69:404–410. doi: 10.1007/s00253-005-1993-3. [DOI] [PubMed] [Google Scholar]
- Kadar Z, De Vrije T, Van Noorden GE, Budde MAW, Szengyel Z, Reczey K, Claassen PAM. Yields from glucose, xylose, and paper sludge hydrolysate during hydrogen production by the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Biochem Biotechnol. 2004;113-116:497–508. doi: 10.1385/abab:114:1-3:497. [DOI] [PubMed] [Google Scholar]
- Lo Y-C, Bai M-D, Chen W-M, Chang J-S. Cellulosic hydrogen production with a sequencing bacterial hydrolysis and dark fermentation strategy. Bioresource Technol. 2008;99:8299–8303. doi: 10.1016/j.biortech.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Ogino H, Miura T, Ishimi K, Seki M, Yoshida H. Hydrogen Production from Glucose by Anaerobes. Biotechnol Prog. 2005;21:1786–1788. doi: 10.1021/bp050224r. [DOI] [PubMed] [Google Scholar]
- Pan C-M, Fan Y-T, Zhao P, Hou H-W. Fermentative hydrogen production by the newly isolated Clostridium beijerinckii Fanp3. Int J Hydrogen Energ. 2008;33:5383–5391. doi: 10.1016/j.ijhydene.2008.05.037. [DOI] [Google Scholar]
- Guo W-Q, Ren N-Q, Wang X-J, Xiang W-S, Ding J, You Y, Liu B-F. Optimization of culture conditions for hydrogen production by Ethanoligenens harbinense B49 using response surface methodology. Bioresource Technol. 2008;100:1192–1196. doi: 10.1016/j.biortech.2008.07.070. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Hallenbeck PC. Response surface methodology for process parameter optimization of hydrogen yield by the metabolically engineered strain Escherichia coli DJT135. Bioresource Technol. 2010;101:1820–1825. doi: 10.1016/j.biortech.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Long C-N, Cui J-J, Liu Z-T, Liu Y-T, Long M-N, Hu Z. Statistical optimization of fermentative hydrogen production from xylose by newly isolated Enterobacter sp. CN1. Int J Hydrogen Energ. 2010;35:6657–6664. doi: 10.1016/j.ijhydene.2010.04.094. [DOI] [Google Scholar]
- Jo JH, Lee DS, Park D, Choe W-S, Park JM. Optimization of key process variables for enhanced hydrogen production by Enterobacter aerogenes using statistical methods. Bioresource Technol. 2008;99:2061–2066. doi: 10.1016/j.biortech.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Wang J, Wan W. Optimization of fermentative hydrogen production process by response surface methodology. Int J Hydrogen Energ. 2008;33:6976–6984. doi: 10.1016/j.ijhydene.2008.08.051. [DOI] [Google Scholar]
- Hallenbeck PC, Ghosh D. Advances in fermentative biohydrogen production: the way forward? Trends Biotechnol. 2009;27:287–297. doi: 10.1016/j.tibtech.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Mu Y, Wang G, Yu H-Q. Response surface methodological analysis on biohydrogen production by enriched anaerobic cultures. Enzyme Microb Technol. 2006;38:905–913. doi: 10.1016/j.enzmictec.2005.08.016. [DOI] [Google Scholar]
- Kuhn M, Steinbuechel A, Schlegel HG. Hydrogen evolution by strictly aerobic hydrogen bacteria under anaerobic conditions. J Bacteriol. 1984;159:633–639. doi: 10.1128/jb.159.2.633-639.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnova ES, Zhilina TN, Tourova TP, Kostrikina NA, Zavarzin GA. Anaerobic, alkaliphilic, saccharolytic bacterium Alkalibacter saccharofermentans gen. nov., sp. nov. from a soda lake in the Transbaikal region of Russia. Extremophiles. 2004;8:309–316. doi: 10.1007/s00792-004-0390-7. [DOI] [PubMed] [Google Scholar]
- Adams CJ, Redmond MC, Valentine DL. Pure-culture growth of fermentative bacteria, facilitated by H2 removal: bioenergetics and H2 production. Appl Environ Microb. 2006;72:1079–1085. doi: 10.1128/AEM.72.2.1079-1085.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle M, Li Y, Woese C, Wiegel J. Isolation and characterization of a novel alkalitolerant thermophile, Anaerobranca horikoshii gen. nov., sp. nov. Int J Syst Bacteriol. 1995;45:454–461. doi: 10.1099/00207713-45-3-454. [DOI] [PubMed] [Google Scholar]
- Strömpl C, Tindall BJ, Jarvis GN, Lunsdorf H, Moore ERB, Hippe H. A re-evaluation of the taxonomy of the genus Anaerovibrio, with the reclassification of Anaerovibrio glycerini as Anaerosinus glycerini gen. nov., comb. nov., and Anaerovibrio burkinabensis as Anaeroarcus burkinensis [corrig.] gen. nov., comb. nov. Int J Syst Bacteriol. 1999;49:1861–1872. doi: 10.1099/00207713-49-4-1861. [DOI] [PubMed] [Google Scholar]
- Prins RA, Lankhorst A, van der Meer P, Van Nevel CJ. Some characteristics of Anaerovibrio lipolytica a rumen lipolytic organism. A van Leeuw J Microb. 1975;41:1–11. doi: 10.1007/BF02565031. [DOI] [PubMed] [Google Scholar]
- Kalia VC, Jain SR, Kumar A, Joshi AP. Fermentation of biowaste to H2 by Bacillus licheniformis. World J Microb Biot. 1994;10:224–227. doi: 10.1007/BF00360893. [DOI] [PubMed] [Google Scholar]
- Sonakya V, Raizada N, Kalia VC. Microbial and enzymatic improvement of anaerobic digestion of waste biomass. Biotechnol Lett. 2001;23:1463–1466. doi: 10.1023/A:1011664912970. [DOI] [Google Scholar]
- Chen M, Wolin MJ. Influence of heme and vitamin B12 on growth and fermentations of Bacteroides species. J Bacteriol. 1981;145:466–471. doi: 10.1128/jb.145.1.466-471.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyenwoke RU, Lee Y-J, Dabrowski S, Ahring BK, Wiegel J. Reclassification of Thermoanaerobium acetigenum as Caldicellulosiruptor acetigenus comb. nov. and emendation of the genus description. Int J Syst Evol Microbiol. 2006;56:1391–1395. doi: 10.1099/ijs.0.63723-0. [DOI] [PubMed] [Google Scholar]
- Bredholt S, Sonne-Hansen J, Nielsen P, Mathrani IM, Ahring BK. Caldicellulosiruptor kristjanssonii sp. nov., a cellulolytic, extremely thermophilic, anaerobic bacterium. Int J Syst Bacteriol. 1999;49:991–996. doi: 10.1099/00207713-49-3-991. [DOI] [PubMed] [Google Scholar]
- Huang CY, Patel BK, Mah RA, Baresi L. Caldicellulosiruptor owensensis sp. nov., an anaerobic, extremely thermophilic, xylanolytic bacterium. Int J Syst Bacteriol. 1998;48(1):91–97. doi: 10.1099/00207713-48-1-91. [DOI] [PubMed] [Google Scholar]
- van Niel EWJ, Claassen PAM, Stams AJM. Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol Bioeng. 2003;81:255–262. doi: 10.1002/bit.10463. [DOI] [PubMed] [Google Scholar]
- Panagiotopoulos IA, Bakker RR, Budde MAW, de Vrije T, Claassen PAM, Koukios EG. Fermentative hydrogen production from pretreated biomass: A comparative study. Bioresource Technol. 2009;100:6331–6338. doi: 10.1016/j.biortech.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Kadar Z, De Vrije T, Budde MAW, Szengyel Z, Reczey K, Claassen PAM. Hydrogen production from paper sludge hydrolysate. Appl Biochem Biotechnol. 2003;105-108:557–566. doi: 10.1385/abab:107:1-3:557. [DOI] [PubMed] [Google Scholar]
- Kanayama H, Sode K, Karube I. Basic studies of hydrogen evolution by Escherichia coli containing a cloned Citrobacter freundii hydrogenase gene. Appl Biochem Biotechnol. 1987;15:97–106. doi: 10.1007/BF02801311. [DOI] [PubMed] [Google Scholar]
- Datta R, Zeikus JG. Modulation of acetone-butanol-ethanol fermentation by carbon monoxide and organic acids. Appl Environ Microbiol. 1985;49:522–529. doi: 10.1128/aem.49.3.522-529.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman KS, Rainey FA, Moe WM. Production of hydrogen by Clostridium species in the presence of chlorinated solvents. FEMS Microbiol Lett. 2009;290:188–194. doi: 10.1111/j.1574-6968.2008.01419.x. [DOI] [PubMed] [Google Scholar]
- Yang JC, Chynoweth DP, Williams DS, Li A. Clostridium aldrichii sp. nov., a cellulolytic mesophile inhabiting a wood-fermenting anaerobic digester. Int J Syst Bacteriol. 1990;40:268–272. doi: 10.1099/00207713-40-3-268. [DOI] [PubMed] [Google Scholar]
- Shcherbakova VA, Chuvilskaya NA, Rivkina EM, Pecheritsyna SA, Laurinavichius KS, Suzina NE, Osipov GA, Lysenko AM, Gilichinsky DA, Akimenko VK. Novel psychrophilic anaerobic spore-forming bacterium from the overcooled water brine in permafrost: description Clostridium algoriphilum sp. nov. Extremophiles. 2005;9:239–246. doi: 10.1007/s00792-005-0438-3. [DOI] [PubMed] [Google Scholar]
- Jayasinghearachchi HS, Singh S, Sarma PM, Aginihotri A, Lal B. Fermentative hydrogen production by new marine Clostridium amygdalinum strain C9 isolated from offshore crude oil pipeline. Int J Hydrogen Energ. 2010;35:6665–6673. doi: 10.1016/j.ijhydene.2010.04.034. [DOI] [Google Scholar]
- Hatch JL, Finneran KT. Influence of reduced electron shuttling compounds on biological H2 production in the fermentative pure culture Clostridium beijerinckii. Curr Microbiol. 2008;56:268–273. doi: 10.1007/s00284-007-9073-9. [DOI] [PubMed] [Google Scholar]
- Wang CC, Chang CW, Chu CP, Lee DJ, Chang BV, Liao CS. Hydrogen Production from Wastewater Sludge Using a Clostridium Strain. J Environ Sci Health Part A Toxic/Hazard Subst Environ Eng. 2003;A38:1867–1875. doi: 10.1081/ese-120022885. [DOI] [PubMed] [Google Scholar]
- Wang CC, Chang CW, Chu CP, Lee DJ, Chang BV, Liao CS. Producing hydrogen from wastewater sludge by Clostridium bifermentans. J Biotechnol. 2003;102:83–92. doi: 10.1016/S0168-1656(03)00007-5. [DOI] [PubMed] [Google Scholar]
- Li D, Chen H. Biological hydrogen production from steam-exploded straw by simultaneous saccharification and fermentation. Int J Hydrogen Energ. 2007;32:1742–1748. doi: 10.1016/j.ijhydene.2006.12.011. [DOI] [Google Scholar]
- Chen W-M, Tseng Z-J, Lee K-S, Chang J-S. Fermentative hydrogen production with Clostridium butyricum CGS5 isolated from anaerobic sewage sludge. Int J Hydrogen Energ. 2005;30:1063–1070. doi: 10.1016/j.ijhydene.2004.09.008. [DOI] [Google Scholar]
- Wang M-Y, Olson BH, Chang J-S. Relationship among growth parameters for Clostridium butyricum, hydA gene expression, and biohydrogen production in a sucrose-supplemented batch reactor. Appl Microbiol Biot. 2008;78:525–532. doi: 10.1007/s00253-007-1317-x. [DOI] [PubMed] [Google Scholar]
- Saratale GD, Saratale RG, Lo Y-C, Chang J-S. Multicomponent cellulase production by Cellulomonas biazotea NCIM-2550 and its applications for cellulosic biohydrogen production. Biotechnol Prog. 2010;26:406–416. doi: 10.1002/btpr.342. [DOI] [PubMed] [Google Scholar]
- Solomon BO, Zeng AP, Biebl H, Schlieker H, Posten C, Deckwer WD. Comparison of the energetic efficiencies of hydrogen and oxychemicals formation in Klebsiella pneumoniae and Clostridium butyricum during anaerobic growth on glycerol. J Biotechnol. 1995;39:107–117. doi: 10.1016/0168-1656(94)00148-6. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wakayama T, Asada Y, Miyake J. Hydrogen production by four cultures with participation by anoxygenic phototrophic bacterium and anaerobic bacteria in the presence of NH4+ Int J Hydrogen Energ. 2001;26:1149–1154. doi: 10.1016/S0360-3199(01)00038-6. [DOI] [Google Scholar]
- Karube I, Matsunaga T, Tsuru S, Suzuki S. Continuous hydrogen production by immobilized whole cells of Clostridium butyricum. Biochim Biophys Acta, Gen Subj. 1976;444:338–343. doi: 10.1016/0304-4165(76)90376-7. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Karube I, Matsunaga T, Kuriyama S, Suzuki N, Shirogami T, Takamura T. Biochemical energy conversion using immobilized whole cells of Clostridium butyricum. Biochimie. 1980;62:353–358. doi: 10.1016/S0300-9084(80)80165-9. [DOI] [PubMed] [Google Scholar]
- Yokoi H, Tokushige T, Hirose J, Hayashi S, Takasaki Y. H2 production from starch by a mixed culture of Clostridium butyricum and Enterobacter aerogenes. Biotechnol Lett. 1998;20:143–147. doi: 10.1023/A:1005372323248. [DOI] [Google Scholar]
- Van Andel JG, Zoutberg GR, Crabbendam PM, Breure AM. Glucose fermentation by Clostridium butyricum grown under a self-generated gas atmosphere in chemostat culture. Appl Microbiol Biot. 1985;23:21–26. doi: 10.1007/BF02660113. [DOI] [Google Scholar]
- Heyndrickx M, De Vos P, Vancanneyt M, De Ley J. The fermentation of glycerol by Clostridium butyricum LMG 1212 t2 and C. pasteurianum LMG 3285. Appl Microbiol Biot. 1991;34:637–642. doi: 10.1007/BF00167914. [DOI] [Google Scholar]
- Heyndrickx M, De Vos P, Speybrouck A, De Ley J. Fermentation of mannitol by Clostridium butyricum: role of acetate as an external hydrogen acceptor. Appl Microbiol Biot. 1989;31:323–328. [Google Scholar]
- Kim M-S, Baek J-S, Yun Y-S, Sim SJ, Park S, Kim S-C. Hydrogen production from Chlamydomonas reinhardtii biomass using a two-step conversion process: Anaerobic conversion and photosynthetic fermentation. Int J Hydrogen Energ. 2006;31:812–816. doi: 10.1016/j.ijhydene.2005.06.009. [DOI] [Google Scholar]
- Brisbarre N, Fardeau M-L, Cueff V, Cayol J-L, Barbier G, Cilia V, Ravot G, Thomas P, Garcia J-L, Ollivier B. Clostridium caminithermale sp. nov., a slightly halophilic and moderately thermophilic bacterium isolated from an Atlantic deep-sea hydrothermal chimney. Int J Syst Evol Microbiol. 2003;53:1043–1049. doi: 10.1099/ijs.0.02471-0. [DOI] [PubMed] [Google Scholar]
- Hungate RE. Studies on cellulose fermentation. I. The culture physiology of an anaerobic cellulose-digesting bacterium. J Bacteriol. 1944;48:499–513. doi: 10.1128/jb.48.5.499-513.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehin A, Cailliez C, Petitdemange E, Benoit L. Studies of Clostridium cellulolyticum ATCC 35319 under dialysis and co-culture conditions. Lett Appl Microbiol. 1996;23:208–212. doi: 10.1111/j.1472-765X.1996.tb00067.x. [DOI] [PubMed] [Google Scholar]
- Doerner C, Schink B. Clostridium homopropionicum sp. nov., a new strict anaerobe growing with 2-, 3-, or 4-hydroxybutyrate. Arch Microbiol. 1990;154:342–348. doi: 10.1007/BF00276529. [DOI] [PubMed] [Google Scholar]
- Monserrate E, Leschine SB, Canale-Parola E. Clostridium hungatei sp. nov., a mesophilic, N2-fixing cellulolytic bacterium isolated from soil. Int J Syst Evol Microbiol. 2001;51:123–132. doi: 10.1099/00207713-51-1-123. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Henninger H, Wenning J, Decker K. Energy metabolism of Clostridium kluyveri. Eur J Biochem. 1968;4:173–180. doi: 10.1111/j.1432-1033.1968.tb00189.x. [DOI] [PubMed] [Google Scholar]
- Varel VH. Reisolation and characterization of Clostridium longisporum, a ruminal sporeforming cellulolytic anaerobe. Arch Microbiol. 1989;152:209–214. doi: 10.1007/BF00409652. [DOI] [PubMed] [Google Scholar]
- Oren A. Clostridium lortetii sp. nov., a halophilic obligatory anaerobic bacterium producing endospores with attached gas vacuoles. Arch Microbiol. 1983;136:42–48. doi: 10.1007/BF00415608. [DOI] [Google Scholar]
- Mechichi T, Labat M, Patel BKC, Woo THS, Thomas P, Garcia J-L. Clostridium methoxybenzovorans sp. nov., a new aromatic o-demethylating homoacetogen from an olive mill wastewater treatment digester. Int J Syst Bacteriol. 1999;49:1201–1209. doi: 10.1099/00207713-49-3-1201. [DOI] [PubMed] [Google Scholar]
- Himelbloom BH, Canale-Parola E. Clostridium methylpentosum sp. nov.: a ring-shaped intestinal bacterium that ferments only methylpentoses and pentoses. Arch Microbiol. 1989;151:287–293. doi: 10.1007/BF00406553. [DOI] [PubMed] [Google Scholar]
- Mechichi T, Fardeau M-L, Labat M, Garcia J-L, Verhe F, Patel BKC. Clostridium peptidivorans sp. nov., a peptide-fermenting bacterium from an olive mill wastewater treatment digester. Int J Syst Evol Microbiol. 2000;50:1259–1264. doi: 10.1099/00207713-50-3-1259. [DOI] [PubMed] [Google Scholar]
- Warnick TA, Methe BA, Leschine SB. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Microbiol. 2002;52:1155–1160. doi: 10.1099/ijs.0.02125-0. [DOI] [PubMed] [Google Scholar]
- Park HS, Kim BH, Kim HS, Kim HJ, Kim GT, Kim M, Chang IS, Park YK, Chang HI. A novel electrochemically active and Fe(III)-reducing bacterium phylogenetically related to Clostridium butyricum isolated from a microbial fuel cell. Anaerobe. 2001;7:297–306. doi: 10.1006/anae.2001.0399. [DOI] [Google Scholar]
- Lamed RJ, Lobos JH, Su TM. Effects of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl Environ Microb. 1988;54:1216–1221. doi: 10.1128/aem.54.5.1216-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling R, Risbey D, Poggi-Varaldo HM. Hydrogen production from inhibited anaerobic composters. Int J Hydrogen Energ. 1997;22:563–566. doi: 10.1016/S0360-3199(96)00137-1. [DOI] [Google Scholar]
- Liu Y, Yu P, Song X, Qu Y. Hydrogen production from cellulose by co-culture of Clostridium thermocellum JN4 and Thermoanaerobacterium thermosaccharolyticum GD17. Int J Hydrogen Energ. 2008;33:2927–2933. doi: 10.1016/j.ijhydene.2008.04.004. [DOI] [Google Scholar]
- Bothun GD, Knutson BL, Berberich JA, Strobel HJ, Nokes SE. Metabolic selectivity and growth of Clostridium thermocellum in continuous culture under elevated hydrostatic pressure. Appl Microbiol Biot. 2004;65:149–157. doi: 10.1007/s00253-004-1554-1. [DOI] [PubMed] [Google Scholar]
- Ng TK, Ben-Bassat A, Zeikus JG. Ethanol production by thermophilic bacteria: fermentation of cellulosic substrates by cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microb. 1981;41:1337–1343. doi: 10.1128/aem.41.6.1337-1343.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J, Ljungdahl LG, Rawson JR. Isolation from soil and properties of the extreme thermophile Clostridium thermohydrosulfuricum. J Bacteriol. 1979;139:800–810. doi: 10.1128/jb.139.3.800-810.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yang S-T. Effect of pH on metabolic pathway shift in fermentation of xylose by Clostridium tyrobutyricum. J Biotechnol. 2004;110:143–157. doi: 10.1016/j.jbiotec.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Mechichi T, Labat M, Garcia JL, Thomas P, Patel BKC. Characterization of a new xylanolytic bacterium, Clostridium xylanovorans sp. nov. Syst Appl Microbiol. 1999;22:366–371. doi: 10.1016/S0723-2020(99)80044-7. [DOI] [PubMed] [Google Scholar]
- Etchebehere C, Pavan ME, Zorzopulos J, Soubes M, Muxi L. Coprothermobacter platensis sp. nov., a new anaerobic proteolytic thermophilic bacterium isolated from an anaerobic mesophilic sludge. Int J Syst Bacteriol. 1998;48:1297–1304. doi: 10.1099/00207713-48-4-1297. [DOI] [PubMed] [Google Scholar]
- Lupton FS, Conrad R, Zeikus JG. Physiological function of hydrogen metabolism during growth of sulfidogenic bacteria on organic substrates. J Bacteriol. 1984;159:843–849. doi: 10.1128/jb.159.3.843-849.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J Bacteriol. 2002;184:5903–5911. doi: 10.1128/JB.184.21.5903-5911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkov AV, Dubinina GA, Lysenko AM, Glockner FO, Kuever J. Dethiosulfovibrio russensis sp. nov., Dethiosulfovibrio marinus sp. nov. and Dethiosulfovibrio acidaminovorans sp. nov., novel anaerobic, thiosulfate- and sulfur-reducing bacteria isolated from "Thiodendron" sulfur mats in different saline environments. Int J Syst Evol Microbiol. 2001;51:327–337. doi: 10.1099/00207713-51-2-327. [DOI] [PubMed] [Google Scholar]
- Trchounian A, Bagramyan K, Poladian A. Formate hydrogenlyase is needed for proton-potassium exchange through the F0F1-ATPase and the TrkA system in anaerobically grown and glycolyzing Escherichia coli. Curr Microbiol. 1997;35:201–206. doi: 10.1007/s002849900239. [DOI] [PubMed] [Google Scholar]
- Penfold DW, Sargent F, Macaskie LE. Inactivation of the Escherichia coli K-12 twin-arginine translocation system promotes increased hydrogen production. FEMS Microbiol Lett. 2006;262:135–137. doi: 10.1111/j.1574-6968.2006.00333.x. [DOI] [PubMed] [Google Scholar]
- Agapakis CM, Ducat DC, Boyle PM, Wintermute EH, Way JC, Silver PA. Insulation of a synthetic hydrogen metabolism circuit in bacteria. J Biol Eng. 2010;4:3. doi: 10.1186/1754-1611-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MA, Mercer J, Mott RA, Pereira-Medrano AG, Burja AM, Radianingtyas H, Wright PC. Engineering a non-native hydrogen production pathway into Escherichia coli via a cyanobacterial [NiFe] hydrogenase. Metab Eng. 2011;13:445–453. doi: 10.1016/j.ymben.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Podesta JJ, Gutierrez-Navarro AM, Estrella CN, Esteso MA. Electrochemical measurement of trace concentrations of biological hydrogen produced by Enterobacteriaceae. Res Microbiol. 1997;148:87–93. doi: 10.1016/S0923-2508(97)81904-3. [DOI] [PubMed] [Google Scholar]
- Stickland LH. The bacterial decomposition of formic acid. Biochem J. 1929;23:1187–1198. doi: 10.1042/bj0231187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnatsakanyan N, Bagramyan K, Trchounian A. Hydrogenase 3 but not hydrogenase 4 is major in hydrogen gas production by Escherichia coli formate hydrogenlyase at acidic pH and in the presence of external formate. Cell Biochem Biophys. 2004;41:357–365. doi: 10.1385/CBB:41:3:357. [DOI] [PubMed] [Google Scholar]
- Bagramyan K, Mnatsakanyan N, Poladian A, Vassilian A, Trchounian A. The roles of hydrogenases 3 and 4, and the F0F1-ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett. 2002;516:172–178. doi: 10.1016/S0014-5793(02)02555-3. [DOI] [PubMed] [Google Scholar]
- Trchounian A, Ohanjanyan Y, Bagramyan K, Vardanian V, Zakharyan E, Vassilian A, Davtian M. Relationship of the Escherichia coli TrkA system of potassium ion uptake with the F0F1-ATPase under growth conditions without anaerobic or aerobic respiration. Biosci Rep. 1998;18:143–154. doi: 10.1023/A:1020144628839. [DOI] [PubMed] [Google Scholar]
- Nandi R, Bhattacharyya PK, Bhaduri AN, Sengupta S. Synthesis and lysis of formate by immobilized cells of Escherichia coli. Biotechnol Bioeng. 1992;39:775–780. doi: 10.1002/bit.260390710. [DOI] [PubMed] [Google Scholar]
- Hatchikian EC, Forget N, Bernadac A, Alazard D, Ollivier B. Involvement of a single periplasmic hydrogenase for both hydrogen uptake and production in some Desulfovibrio species. Res Microbiol. 1995;146:129–141. doi: 10.1016/0923-2508(96)80891-6. [DOI] [PubMed] [Google Scholar]
- Xing D, Ren N, Li Q, Lin M, Wang A, Zhao L. Ethanoligenens harbinense gen. nov., sp. nov., isolated from molasses wastewater. Int J Syst Evol Microbiol. 2006;56:755–760. doi: 10.1099/ijs.0.63926-0. [DOI] [PubMed] [Google Scholar]
- Andrews KT, Patel BK. Fervidobacterium gondwanense sp. nov., a new thermophilic anaerobic bacterium isolated from nonvolcanically heated geothermal waters of the Great Artesian Basin of Australia. Int J Syst Bacteriol. 1996;46:265–269. doi: 10.1099/00207713-46-1-265. [DOI] [PubMed] [Google Scholar]
- van Ooteghem SA, Jones A, Van der Lelie D, Dong B, Mahajan D. H2 production and carbon utilization by Thermotoga neapolitana under anaerobic and microaerobic growth conditions. Biotechnol Lett. 2004;26:1223–1232. doi: 10.1023/B:BILE.0000036602.75427.88. [DOI] [PubMed] [Google Scholar]
- Cord-Ruwisch R, Lovley DR, Schink B. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl Environ Microb. 1998;64:2232–2236. doi: 10.1128/aem.64.6.2232-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiret FG, Ortega E, Kleiner D, Ortega-Rodes P, Rodes R, Dong Z. A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J Appl Microbiol. 2004;97:504–511. doi: 10.1111/j.1365-2672.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- Brown SD, Begemann MB, Mormile MR, Wall JD, Han CS, Goodwin LA, Pitluck S, Land ML, Hauser LJ, Elias DA. Complete genome sequence of the haloalkaliphilic, hydrogen-producing bacterium Halanaerobium hydrogeniformans. J Bacteriol. 2011;193:3682–3683. doi: 10.1128/JB.05209-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayol JL, Ollivier B, Patel BK, Ageron E, Grimont PA, Prensier G, Garcia JL. Haloanaerobium lacusroseus sp. nov., an extremely halophilic fermentative bacterium from the sediments of a hypersaline lake. Int J Syst Bacteriol. 1995;45:790–797. doi: 10.1099/00207713-45-4-790. [DOI] [PubMed] [Google Scholar]
- Cayol J-L, Ollivier B, Lawson ASA, Fardeau M-L, Ageron E, Grimont PAD, Prensier G, Guezennec J, Magot M, Garcia JL. Haloincola saccharolytica subsp. senegalensis subsp. nov., isolated from the sediments of a hypersaline lake, and emended description of Haloincola saccharolytica. Int J Syst Bacteriol. 1994;44:805–811. doi: 10.1099/00207713-44-4-805. [DOI] [Google Scholar]
- Cayol JL, Ollivier B, Patel BKC, Prensier G, Guezennec J, Garcia JL. Isolation and characterization of Halothermothrix orenii gen. nov., sp. nov., a halophilic, thermophilic, fermentative, strictly anaerobic bacterium. Int J Syst Bacteriol. 1994;44:534–540. doi: 10.1099/00207713-44-3-534. [DOI] [PubMed] [Google Scholar]
- Brune A, Evers S, Kaim G, Ludwig W, Schink B. Ilyobacter insuetus sp. nov., a fermentative bacterium specialized in the degradation of hydroaromatic compounds. Int J Syst Evol Microbiol. 2002;52:429–432. doi: 10.1099/00207713-52-2-429. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Q, Dieudonne M, Cong Y, Zhou J, Long M. Enhanced H2 gas production from bagasse using adhE inactivated Klebsiella oxytoca HP1 by sequential dark-photo fermentations. Bioresource Technol. 2010;101:9605–9611. doi: 10.1016/j.biortech.2010.07.095. [DOI] [PubMed] [Google Scholar]
- Niu K, Zhang X, Tan W-S, Zhu M-L. Effect of culture conditions on producing and uptake hydrogen flux of biohydrogen fermentation by metabolic flux analysis method. Bioresource Technol. 2011;102:7294–7300. doi: 10.1016/j.biortech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Schink B. Fermentation of 2,3-butanediol by Pelobacter carbinolicus sp. nov. and Pelobacter propionicus sp. nov., and evidence for propionate formation from C2 compounds. Arch Microbiol. 1984;137:33–41. doi: 10.1007/BF00425804. [DOI] [Google Scholar]
- Guo L, Li X-M, Bo X, Yang Q, Zeng G-M, Liao D-x, Liu J-J. Impacts of sterilization, microwave and ultrasonication pretreatment on hydrogen producing using waste sludge. Bioresource Technol. 2008;99:3651–3658. doi: 10.1016/j.biortech.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Chassard C, Bernalier-Donadille A. H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol Lett. 2006;254:116–122. doi: 10.1111/j.1574-6968.2005.00016.x. [DOI] [PubMed] [Google Scholar]
- Hungate RE. Microorganisms in the rumen of cattle fed a constant ration. Can J Microbiol. 1957;3:289–311. doi: 10.1139/m57-034. [DOI] [PubMed] [Google Scholar]
- Kistner A, Gouws L. Cellulolytic cocci occurring in the rumen of sheep conditioned to lucerne hay. J Gen Microbiol. 1964;34:447–458. doi: 10.1099/00221287-34-3-447. [DOI] [PubMed] [Google Scholar]
- Goodwin S, Zeikus JG. Ecophysiological adaptations of anaerobic bacteria to low pH: analysis of anaerobic digestion in acidic bog sediments. Appl Environ Microb. 1987;53:57–64. doi: 10.1128/aem.53.1.57-64.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Plugge CM, Roelofsen W, Houwen FP, Stams AJM. Selenomonas acidaminovorans sp. nov., a versatile thermophilic proton-reducing anaerobe able to grow by decarboxylation of succinate to propionate. Arch Microbiol. 1992;157:169–175. [Google Scholar]
- Scheifinger CC, Linehan B, Wolin MJ. H2 production by Selenomonas ruminantium in the absence and presence of methanogenic bacteria. Appl Microbiol. 1975;29:480–483. doi: 10.1128/am.29.4.480-483.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin MJ. Metabolic interactions among intestinal microorganisms. Am J Clin Nutr. 1974;27:1320–1328. doi: 10.1093/ajcn/27.11.1320. [DOI] [PubMed] [Google Scholar]
- Breznak JA, Canale-Parola E. Morphology and physiology of Spirochaeta aurantia strains isolated from aquatic habitats. Arch Microbiol. 1975;105:1–12. doi: 10.1007/BF00447104. [DOI] [PubMed] [Google Scholar]
- Slobodkin AI, Tourova TP, Kostrikina NA, Chernyh NA, Bonch-Osmolovskaya EA, Jeanthon C, Jones BE. Tepidibacter thalassicus gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, fermentative bacterium from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2003;53:1131–1134. doi: 10.1099/ijs.0.02600-0. [DOI] [PubMed] [Google Scholar]
- Zeikus JG, Hegge PW, Anderson MA. Thermoanaerobium brockii genus nova and species nova, a new chemoorganotrophic, caldoactive, anaerobic bacterium. Arch Microbiol. 1979;122:41–48. doi: 10.1007/BF00408044. [DOI] [Google Scholar]
- Lee Y-E, Jain MK, Lee C, Lowe SE, Zeikus JG. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov. , sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov. ; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfiricum ElO0-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov. , and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int J Syst Bacteriol. 1993;43:41–51. doi: 10.1099/00207713-43-1-41. [DOI] [Google Scholar]
- Cayol JL, Ollivier B, Patel BK, Ravot G, Magot M, Ageron E, Grimont PA, Garcia JL. Description of Thermoanaerobacter brockii subsp. lactiethylicus subsp. nov., isolated from a deep subsurface French oil well, a proposal to reclassify Thermoanaerobacter finnii as Thermoanaerobacter brockii subsp. finnii comb. nov., and an emended description of Thermoanaerobacter brockii. Int J Syst Bacteriol. 1995;45:783–789. doi: 10.1099/00207713-45-4-783. [DOI] [PubMed] [Google Scholar]
- Slobodkin AI, Tourova TP, Kuznetsov BB, Kostrikina NA, Chernyh NA, Bonch-Osmolovskaya EA. Thermoanaerobacter siderophilus sp. nov., a novel dissimilatory Fe(III)-reducing, anaerobic, thermophilic bacterium. Int J Syst Bacteriol. 1999;49(4):1471–1478. doi: 10.1099/00207713-49-4-1471. [DOI] [PubMed] [Google Scholar]
- Kublanov IV, Prokofeva MI, Kostrikina NA, Kolganova TV, Tourova TP, Wiegel J, Bonch-Osmolovskaya EA. Thermoanaerobacterium aciditolerans sp. nov., a moderate thermoacidophile from a Kamchatka hot spring. Int J Syst Evol Microbiol. 2007;57:260–264. doi: 10.1099/ijs.0.64633-0. [DOI] [PubMed] [Google Scholar]
- Cann IK, Stroot PG, Mackie KR, White BA, Mackie RI. Characterization of two novel saccharolytic, anaerobic thermophiles, Thermoanaerobacterium polysaccharolyticum sp. nov. and Thermoanaerobacterium zeae sp. nov., and emendation of the genus Thermoanaerobacterium. Int J Syst Evol Microbiol. 2001;51:293–302. doi: 10.1099/00207713-51-2-293. [DOI] [PubMed] [Google Scholar]
- Ben-Bassat A, Lamed R, Zeikus JG. Ethanol production by thermophilic bacteria: metabolic control of end product formation in Thermoanaerobium brockii. J Bacteriol. 1981;146:192–199. doi: 10.1128/jb.146.1.192-199.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle M, Li Y, Rainey F, DeBlois S, Mai V, Reichert A, Mayer F, Messner P, Wiegel J. Thermobrachium celere gen. nov., sp. nov., a rapidly growing thermophilic, alkalitolerant, and proteolytic obligate anaerobe. Int J Syst Bacteriol. 1996;46:1025–1033. doi: 10.1099/00207713-46-4-1025. [DOI] [PubMed] [Google Scholar]
- Ciranna A, Santala V, Karp M. Biohydrogen production in alkalithermophilic conditions: Thermobrachium celere as a case study. Bioresource Technol. 2011;102:8714–8722. doi: 10.1016/j.biortech.2011.01.028. [DOI] [PubMed] [Google Scholar]
- Ma K, Loessner H, Heider J, Johnson MK, Adams MWW. Effects of elemental sulfur on the metabolism of the deep-sea hyperthermophilic archaeon Thermococcus strain ES-1: characterization of a sulfur-regulated, non-heme iron alcohol dehydrogenase. J Bacteriol. 1995;177:4748–4756. doi: 10.1128/jb.177.16.4748-4756.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrije T, de Haas GG, Tan GB, Keijsers ERP, Claassen PAM. Pretreatment of Miscanthus for hydrogen production by Thermotoga elfii. Int J Hydrogen Energ. 2002;27:1381–1390. doi: 10.1016/S0360-3199(02)00124-6. [DOI] [Google Scholar]
- Balk M, Weijma J, Stams AJM. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int J Syst Evol Microbiol. 2002;52:1361–1368. doi: 10.1099/ijs.0.02165-0. [DOI] [PubMed] [Google Scholar]
- Nguyen TAD, Kim JP, Kim MS, Oh YK, Sim SJ. Optimization of hydrogen production by hyperthermophilic eubacteria, Thermotoga maritima and Thermotoga neapolitana in batch fermentation. Int J Hydrogen Energ. 2008;33:1483–1488. doi: 10.1016/j.ijhydene.2007.09.033. [DOI] [Google Scholar]
- Zavarzina DG, Tourova TP, Kuznetsov BB, Bonch-Osmolovskaya EA, Slobodkin AI. Thermovenabulum ferriorganovorum gen. nov., sp. nov., a novel thermophilic, anaerobic, endospore-forming bacterium. Int J Syst Evol Microbiol. 2002;52:1737–1743. doi: 10.1099/ijs.0.02214-0. [DOI] [PubMed] [Google Scholar]
- Shieh WY, Chen A-L, Chiu H-H. Vibrio aerogenes sp. nov., a facultatively anaerobic marine bacterium that ferments glucose with gas production. Int J Syst Evol Microbiol. 2000;50:321–329. doi: 10.1099/00207713-50-1-321. [DOI] [PubMed] [Google Scholar]
- Wang A, Ren N, Shi Y, Lee D-J. Bioaugmented hydrogen production from microcrystalline cellulose using co-culture-Clostridium acetobutylicum X9 and Ethanoigenens harbinense B49. Int J Hydrogen Energ. 2008;33:912–917. doi: 10.1016/j.ijhydene.2007.10.017. [DOI] [Google Scholar]
- Kodama Y, Watanabe K. An electricity-generating prosthecate bacterium strain Mfc52 isolated from a microbial fuel cell. FEMS Microbiol Lett. 2008;288:55–61. doi: 10.1111/j.1574-6968.2008.01326.x. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Cox D, Levandowsky M. Production of hydrogen by microbial fermentation. Int J Hydrogen Energ. 1988;13:407–410. doi: 10.1016/0360-3199(88)90126-7. [DOI] [Google Scholar]
- Geng A, He Y, Qian C, Yan X, Zhou Z. Effect of key factors on hydrogen production from cellulose in a co-culture of Clostridium thermocellum and Clostridium thermopalmarium. Bioresource Technol. 2010;101:4029–4033. doi: 10.1016/j.biortech.2010.01.042. [DOI] [PubMed] [Google Scholar]
- Diekert G, Ritter M. Nickel requirement of Acetobacterium woodii. J Bacteriol. 1982;151:1043–1045. doi: 10.1128/jb.151.2.1043-1045.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GM, Russell JB. Dual mechanisms of tricarboxylate transport and catabolism by Acidaminococcus fermentans. Appl Environ Microb. 1994;60:2538–2544. doi: 10.1128/aem.60.7.2538-2544.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haertel U, Buckel W. Sodium ion-dependent hydrogen production in Acidaminococcus fermentans. Arch Microbiol. 1996;166:350–356. doi: 10.1007/s002030050394. [DOI] [PubMed] [Google Scholar]
- Haertel U, Buckel W. Fermentation of trans-aconitate via citrate, oxaloacetate, and pyruvate by Acidaminococcus fermentans. Arch Microbiol. 1996;166:342–349. doi: 10.1007/s002030050393. [DOI] [PubMed] [Google Scholar]
- Koku H, Eroglu I, Gunduz U, Yucel M, Turker L. Aspects of the metabolism of hydrogen production by Rhodobacter sphaeroides. Int J Hydrogen Energ. 2002;27:1315–1329. doi: 10.1016/S0360-3199(02)00127-1. [DOI] [Google Scholar]
- Seyfried M, Lyon D, Rainey FA, Wiegel J. Caloramator viterbensis sp. nov., a novel thermophilic, glycerol-fermenting bacterium isolated from a hot spring in Italy. Int J Syst Evol Microbiol. 2002;52:1177–1184. doi: 10.1099/ijs.0.01472-0. [DOI] [PubMed] [Google Scholar]
- Oh Y-K, Kim H-J, Park S, Kim M-S, Ryu DDY. Metabolic-flux analysis of hydrogen production pathway in Citrobacter amalonaticus Y19. Int J Hydrogen Energ. 2008;33:1471–1482. doi: 10.1016/j.ijhydene.2007.09.032. [DOI] [Google Scholar]
- Kim S, Seol E, Mohan Raj S, Park S, Oh Y-K, Ryu DDY. Various hydrogenases and formate-dependent hydrogen production in Citrobacter amalonaticus Y19. Int J Hydrogen Energ. 2008;33:1509–1515. doi: 10.1016/j.ijhydene.2007.09.029. [DOI] [Google Scholar]
- Oh Y-K, Seol E-H, Kim JR, Park S. Fermentative biohydrogen production by a new chemoheterotrophic bacterium Citrobacter sp. Y19. Int J Hydrogen Energ. 2003;28:1353–1359. doi: 10.1016/S0360-3199(03)00024-7. [DOI] [Google Scholar]
- Kim BH, Bellows P, Datta R, Zeikus JG. Control of carbon and electron flow in Clostridium acetobutylicum fermentations: utilization of carbon monoxide to inhibit hydrogen production and to enhance butanol yields. Appl Environ Microb. 1984;48:764–770. doi: 10.1128/aem.48.4.764-770.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Ward TE, Logan BE, Regan JM. Characterization of the cellulolytic and hydrogen-producing activities of six mesophilic Clostridium species. J Appl Microbiol. 2007;103:2258–2266. doi: 10.1111/j.1365-2672.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- Alshiyab H, Kalil MS, Hamid AA, Yusoff WMW. Removal of headspace CO2 increases biological hydrogen production by C. acetobutylicum. Pak J Biol Sci. 2008;11:2336–2340. doi: 10.3923/pjbs.2008.2336.2340. [DOI] [PubMed] [Google Scholar]
- Jeong T-Y, Cha G-C, Yeom SH, Choi SS. Comparison of hydrogen production by four representative hydrogen-producing bacteria. J Ind Eng. 2008;14:333–337. doi: 10.1016/j.jiec.2007.09.014. [DOI] [Google Scholar]
- Zhao X, Xing D, Fu N, Liu B, Ren N. Hydrogen production by the newly isolated Clostridium beijerinckii RZF-1108. Bioresource Technol. 2011;102:8432–8436. doi: 10.1016/j.biortech.2011.02.086. [DOI] [PubMed] [Google Scholar]
- Lo Y-C, Chen W-M, Hung C-H, Chen S-D, Chang J-S. Dark H2 fermentation from sucrose and xylose using H2-producing indigenous bacteria: Feasibility and kinetic studies. Water Res. 2008;42:827–842. doi: 10.1016/j.watres.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Chen S-D, Sheu D-S, Chen W-M, Lo Y-C, Huang T-I, Lin C-Y, Chang J-S. Dark Hydrogen Fermentation from Hydrolyzed Starch Treated with Recombinant Amylase Originating from Caldimonas taiwanensis On1. Biotechnol Prog. 2007;23:1312–1320. doi: 10.1021/bp070187z. [DOI] [PubMed] [Google Scholar]
- Lo Y-C, Huang L-F, Cheng C-L, Chen J, Chang J-S. Using a starch-rich mutant of Arabidopsis thaliana as feedstock for fermentative hydrogen production. Bioresource Technol. 2011;102:8543–8546. doi: 10.1016/j.biortech.2011.04.034. [DOI] [PubMed] [Google Scholar]
- Lo Y-C, Lu W-C, Chen C-Y, Chang J-S. Dark fermentative hydrogen production from enzymatic hydrolysate of xylan and pretreated rice straw by Clostridium butyricum CGS5. Bioresource Technol. 2010;101:5885–5891. doi: 10.1016/j.biortech.2010.02.085. [DOI] [PubMed] [Google Scholar]
- Lo Y-C, Su Y-C, Chen C-Y, Chen W-M, Lee K-S, Chang J-S. Biohydrogen production from cellulosic hydrolysate produced via temperature-shift-enhanced bacterial cellulose hydrolysis. Bioresource Technol. 2009;100:5802–5807. doi: 10.1016/j.biortech.2009.06.066. [DOI] [PubMed] [Google Scholar]
- Fang HHP, Zhu H, Zhang T. Phototrophic hydrogen production from glucose by pure and co-cultures of Clostridium butyricum and Rhodobacter sphaeroides. Int J Hydrogen Energ. 2006;31:2223–2230. doi: 10.1016/j.ijhydene.2006.03.005. [DOI] [Google Scholar]
- Karube I, Urano N, Matsunaga T, Suzuki S. Hydrogen production from glucose by immobilized growing cells of Clostridium butyricum. Eur J Appl Microbiol Biotechnol. 1982;16:5–9. doi: 10.1007/BF01008235. [DOI] [Google Scholar]
- Yokoi H, Mori S, Hirose J, Hayashi S, Takasaki Y. H2 production from starch by a mixed culture of Clostridium butyricum and Rhodobacter sp. M-19. Biotechnol Lett. 1998;20:895–899. doi: 10.1023/A:1005327912678. [DOI] [Google Scholar]
- Jen CJ, Chou C-H, Hsu P-C, Yu S-J, Chen W-E, Lay J-J, Huang C-C, Wen F-S. Flow-FISH analysis and isolation of clostridial strains in an anaerobic semi-solid bio-hydrogen producing system by hydrogenase gene target. Appl Microbiol Biot. 2007;74:1126–1134. doi: 10.1007/s00253-006-0740-8. [DOI] [PubMed] [Google Scholar]
- Vavilin VA, Rytow SV, Lokshina LY. Modeling hydrogen partial pressure change as a result of competition between the butyric and propionic groups of acidogenic bacteria. Bioresource Technol. 1995;54:171–177. doi: 10.1016/0960-8524(95)00127-1. [DOI] [Google Scholar]
- Pattra S, Sangyoka S, Boonmee M, Reungsang A. Bio-hydrogen production from the fermentation of sugarcane bagasse hydrolysate by Clostridium butyricum. Int J Hydrogen Energ. 2008;33:5256–5265. doi: 10.1016/j.ijhydene.2008.05.008. [DOI] [Google Scholar]
- Ho K-L, Chen Y-Y, Lee D-J. Biohydrogen production from cellobiose in phenol and cresol-containing medium using Clostridium sp. R1. Int J Hydrogen Energ. 2010;35:10239–10244. doi: 10.1016/j.ijhydene.2010.07.155. [DOI] [Google Scholar]
- Liu B-F, Ren N-Q, Xie G-J, Ding J, Guo W-Q, Xing D-F. Enhanced bio-hydrogen production by the combination of dark- and photo-fermentation in batch culture. Bioresource Technol. 2010;101:5325–5329. doi: 10.1016/j.biortech.2010.02.024. [DOI] [PubMed] [Google Scholar]
- Chung KT. Inhibitory effects of H2 on growth of Clostridium cellobioparum. Appl Environ Microbiol. 1976;31:342–348. doi: 10.1128/aem.31.3.342-348.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WJ, Asmundson RV, Hopcroft DH. Isolation and characterization of a strictly anaerobic, cellulolytic spore former: Clostridium chartatabidum sp. nov. Arch Microbiol. 1987;147:169–173. doi: 10.1007/BF00415279. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Nishimura Y. Hydrogen production by fermentation using acetic acid and lactic acid. J Biosci Bioeng. 2007;103:236–241. doi: 10.1263/jbb.103.236. [DOI] [PubMed] [Google Scholar]
- Chen C-Y, Yang M-H, Yeh K-L, Liu C-H, Chang J-S. Biohydrogen production using sequential two-stage dark and photo fermentation processes. Int J Hydrogen Energ. 2008;33:4755–4762. doi: 10.1016/j.ijhydene.2008.06.055. [DOI] [Google Scholar]
- Alalayah WM, Kalil MS, Kadhum AAH, Jahim JM, Jaapar SZS, Alauj NM. Bio-hydrogen production using a two-stage fermentation process. Pak J Biol Sci. 2009;12:1462–1467. doi: 10.3923/pjbs.2009.1462.1467. [DOI] [PubMed] [Google Scholar]
- Ferchichi M, Crabbe E, Hintz W, Gil G-H, Almadidy A. Influence of Culture Parameters on Biological Hydrogen Production by Clostridium saccharoperbutylacetonicum ATCC 27021. World J Microb Biot. 2005;21:855–862. doi: 10.1007/s11274-004-5972-0. [DOI] [Google Scholar]
- Ferchichi M, Crabbe E, Gil G-H, Hintz W, Almadidy A. Influence of initial pH on hydrogen production from cheese whey. J Biotechnol. 2005;120:402–409. doi: 10.1016/j.jbiotec.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Fan Y-T, Xing Y, Ma H-C, Pan C-M, Hou H-W. Enhanced cellulose-hydrogen production from corn stalk by lesser panda manure. Int J Hydrogen Energ. 2008;33:6058–6065. doi: 10.1016/j.ijhydene.2008.08.005. [DOI] [Google Scholar]
- Taguchi F, Mizukami N, Yamada K, Hasegawa K, Saito-Taki T. Direct conversion of cellulosic materials to hydrogen by Clostridium sp. strain no. 2. Enzyme Microb Technol. 1995;17:147–150. doi: 10.1016/0141-0229(94)00058-Y. [DOI] [Google Scholar]
- Taguchi F, Hasegawa K, Saito-Taki T, Hara K. Simultaneous production of xylanase and hydrogen using xylan in batch culture of Clostridium sp. strain X53. J Ferment Bioeng. 1996;81:178–180. doi: 10.1016/0922-338X(96)87600-8. [DOI] [Google Scholar]
- Li Y, Engle M, Weiss N, Mandelco L, Wiegel J. Clostridium thermoalcaliphilum sp. nov., an anaerobic and thermotolerant facultative alkaliphile. Int J Syst Bacteriol. 1994;44:111–118. doi: 10.1099/00207713-44-1-111. [DOI] [PubMed] [Google Scholar]
- Wiegel J, Kuk SU, Kohring GW. Clostridium thermobutyricum sp. nov., a moderate thermophile isolated from a cellulolytic culture, that produces butyrate as the major product. Int J Syst Bacteriol. 1989;39:199–204. doi: 10.1099/00207713-39-2-199. [DOI] [Google Scholar]
- Levin DB, Islam R, Cicek N, Sparling R. Hydrogen production by Clostridium thermocellum 27405 from cellulosic biomass substrates. Int J Hydrogen Energ. 2006;31:1496–1503. doi: 10.1016/j.ijhydene.2006.06.015. [DOI] [Google Scholar]
- Islam R, Cicek N, Sparling R, Levin D. Influence of initial cellulose concentration on the carbon flow distribution during batch fermentation by Clostridium thermocellum ATCC 27405. Appl Microbiol Biot. 2009;82:141–148. doi: 10.1007/s00253-008-1763-0. [DOI] [PubMed] [Google Scholar]
- Islam R, Cicek N, Sparling R, Levin D. Effect of substrate loading on hydrogen production during anaerobic fermentation by Clostridium thermocellum 27405. Appl Microbiol Biot. 2006;72:576–583. doi: 10.1007/s00253-006-0316-7. [DOI] [PubMed] [Google Scholar]
- Magnusson L, Islam R, Sparling R, Levin D, Cicek N. Direct hydrogen production from cellulosic waste materials with a single-step dark fermentation process. Int J Hydrogen Energ. 2008;33:5398–5403. doi: 10.1016/j.ijhydene.2008.06.018. [DOI] [Google Scholar]
- Weimer PJ, Zeikus JG. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol. 1977;33:289–297. doi: 10.1128/aem.33.2.289-297.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun HH, Shen GJ, Zeikus JG. Differential amylosaccharide metabolism of Clostridium thermosulfurogenes and Clostridium thermohydrosulfuricum. J Bacteriol. 1985;164:1153–1161. doi: 10.1128/jb.164.3.1153-1161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovitt RW, Shen GJ, Zeikus JG. Ethanol production by thermophilic bacteria: biochemical basis for ethanol and hydrogen tolerance in Clostridium thermohydrosulfuricum. J Bacteriol. 1988;170:2809–2815. doi: 10.1128/jb.170.6.2809-2815.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talabardon M, Schwitzguebel JP, Peringer P. Anaerobic thermophilic fermentation for acetic acid production from milk permeate. J Biotechnol. 1999;76:83–92. doi: 10.1016/s0168-1656(99)00180-7. [DOI] [PubMed] [Google Scholar]
- Sridhar J, Eiteman MA, Wiegel JW. Elucidation of enzymes in fermentation pathways used by Clostridium thermosuccinogenes growing on inulin. Appl Environ Microb. 2000;66:246–251. doi: 10.1128/AEM.66.1.246-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B, Zeikus JG. Clostridium thermosulfurogenes sp. nov., a new thermophile that produces elemental sulfur from thiosulfate. J Gen Microbiol. 1983;129:1149–1158. [Google Scholar]
- Matthies C, Kuhner CH, Acker G, Drake HL. Clostridium uliginosum sp. nov., a novel acid-tolerant, anaerobic bacterium with connecting filaments. Int J Syst Evol Microbiol. 2001;51:1119–1125. doi: 10.1099/00207713-51-3-1119. [DOI] [PubMed] [Google Scholar]
- Schnurer A, Schink B, Svensson BH. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int J Syst Bacteriol. 1996;46:1145–1152. doi: 10.1099/00207713-46-4-1145. [DOI] [PubMed] [Google Scholar]
- Rachman MA, Furutani Y, Nakashimada Y, Kakizono T, Nishio N. Enhanced hydrogen production in altered mixed acid fermentation of glucose by Enterobacter aerogenes. J Ferment Bioeng. 1997;83:358–363. doi: 10.1016/S0922-338X(97)80142-0. [DOI] [Google Scholar]
- Yokoi H, Ohkawara T, Hirose J, Hayashi S, Takasaki Y. Characteristics of hydrogen production by aciduric Enterobacter aerogenes strain HO-39. J Ferment Bioeng. 1995;80:571–574. doi: 10.1016/0922-338X(96)87733-6. [DOI] [Google Scholar]
- Yokoi H, Tokushige T, Hirose J, Hayashi S, Takasaki Y. Hydrogen production by immobilized cells of aciduric Enterobacter aerogenes strain HO-39. J Ferment Bioeng. 1997;83:481–484. doi: 10.1016/S0922-338X(97)83006-1. [DOI] [Google Scholar]
- Ito T, Nakashimada Y, Senba K, Matsui T, Nishio N. Hydrogen and ethanol production from glycerol-containing wastes discharged after biodiesel manufacturing process. Seibutsu-Kogaku Kais. 2007;85:15. doi: 10.1263/jbb.100.260. [DOI] [PubMed] [Google Scholar]
- Nakashimada Y, Rachman MA, Kakizono T, Nishio N. Hydrogen production of Enterobacter aerogenes altered by extracellular and intracellular redox states. Int J Hydrogen Energ. 2002;27:1399–1405. doi: 10.1016/S0360-3199(02)00128-3. [DOI] [Google Scholar]
- Ito T, Nakashimada Y, Kakizono T, Nishio N. High-yield production of hydrogen by Enterobacter aerogenes mutants with decreased alpha -acetolactate synthase activity. J Biosci Bioeng. 2004;97:227–232. doi: 10.1016/S1389-1723(04)70196-6. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xing X-H, Lou K. Rapid detection of a GFP-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol Lett. 2005;249:211–218. doi: 10.1016/j.femsle.2005.05.051. [DOI] [PubMed] [Google Scholar]
- Tanisho S, Wakao N, Kosako Y. Biological hydrogen production by Enterobacter aerogenes. J Chem Eng Jpn. 1983;16:529–530. doi: 10.1252/jcej.16.529. [DOI] [Google Scholar]
- Potrikus CJ, Breznak JA. Nitrogen-fixing Enterobacter agglomerans isolated from guts of wood-eating termites. Appl Environ Microb. 1977;33:392–399. doi: 10.1128/aem.33.2.392-399.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Fan S-Q, Tian L, Pan C-M, Fan Y-T, Hou H-W. Hydrogen production characteristics from dark fermentation of maltose by an isolated strain F..P 01. Int J Hydrogen Energ. 2010;35:7189–7193. doi: 10.1016/j.ijhydene.2009.12.188. [DOI] [Google Scholar]
- Robert C, Del'Homme C, Bernalier-Donadille A. Interspecies H2 transfer in cellulose degradation between fibrolytic bacteria and H2-utilizing microorganisms from the human colon. FEMS Microbiol Lett. 2001;205:209–214. doi: 10.1111/j.1574-6968.2001.tb10949.x. [DOI] [PubMed] [Google Scholar]
- Akhtar MK, Jones PR. Deletion of iscR stimulates recombinant clostridial Fe-Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3) Appl Microbiol Biot. 2008;78:853–862. doi: 10.1007/s00253-008-1377-6. [DOI] [PubMed] [Google Scholar]
- Kim JYH, Jo BH, Cha HJ. Production of biohydrogen by recombinant expression of [NiFe]-hydrogenase 1 in Escherichia coli. Microb Cell Fact. 2010;9:54. doi: 10.1186/1475-2859-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JYH, Jo BH, Cha HJ. Production of biohydrogen by heterologous expression of oxygen-tolerant Hydrogenovibrio marinus [NiFe]-hydrogenase in Escherichia coli. J Biotechnol. 2011;155:312–319. doi: 10.1016/j.jbiotec.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Kim YM, Cho H-S, Jung GY, Park JM. Engineering the pentose phosphate pathway to improve hydrogen yield in recombinant Escherichia coli. Biotechnol Bioeng. 2011;108:2941–2946. doi: 10.1002/bit.23259. [DOI] [PubMed] [Google Scholar]
- Hu H, Wood TK. An evolved Escherichia coli strain for producing hydrogen and ethanol from glycerol. Biochem Biophys Res Commun. 2010;391:1033–1038. doi: 10.1016/j.bbrc.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Bisaillon A, Turcot J, Hallenbeck PC. The effect of nutrient limitation on hydrogen production by batch cultures of Escherichia coli. Int J Hydrogen Energ. 2006;31:1504–1508. doi: 10.1016/j.ijhydene.2006.06.016. [DOI] [Google Scholar]
- Hill S, Viollet S, Smith AT, Anthony C. Roles for enteric d-type cytochrome oxidase in N2 fixation and microaerobiosis. J Bacteriol. 1990;172:2071–2078. doi: 10.1128/jb.172.4.2071-2078.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Yamamura S, Takamura Y, Sode K, Tamiya E, Tomiyama M. Development of a compact high-density microbial hydrogen reactor for portable bio-fuel cell system. Int J Hydrogen Energ. 2006;31:1484–1489. doi: 10.1016/j.ijhydene.2006.06.014. [DOI] [Google Scholar]
- Sode K, Watanabe M, Makimoto H, Tomiyama M. Construction and characterization of fermentative lactate dehydrogenase Escherichia coli mutant and its potential for bacterial hydrogen production. Appl Biochem Biotechnol. 1999;77-79:317–323. [Google Scholar]
- Tikka J. Über den Mechanismus der Glukosevergährung von B. coli. Biochem Z. 1935;279:264–288. [Google Scholar]
- Fan Z, Yuan L, Chatterjee R. Increased hydrogen production by genetic engineering of Escherichia coli. PLoS One. 2009;4(2):4432. doi: 10.1371/journal.pone.0004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B-F, Ren N-Q, Xing D-F, Ding J, Zheng G-X, Guo W-Q, Xu J-F, Xie G-J. Hydrogen production by immobilized R. faecalis RLD-53 using soluble metabolites from ethanol fermentation bacteria E. harbinense B49. Bioresource Technol. 2009;100:2719–2723. doi: 10.1016/j.biortech.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Xing D, Ren N, Wang A, Li Q, Feng Y, Ma F. Continuous hydrogen production of auto-aggregative Ethanoligenens harbinense YUAN-3 under non-sterile condition. Int J Hydrogen Energ. 2008;33:1489–1495. doi: 10.1016/j.ijhydene.2007.09.038. [DOI] [Google Scholar]
- Ravot G, Ollivier B, Fardeau M-L, Patel BKC, Andrews KT, Magot M, Garcia J-L. L-Alanine production from glucose fermentation by hyperthermophilic members of the domains Bacteria and Archaea: a remnant of an ancestral metabolism? Appl Environ Microb. 1996;62:2657–2659. doi: 10.1128/aem.62.7.2657-2659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravot G, Magot M, Fardeau M-L, Patel BKC, Thomas P, Garcia J-L, Ollivier B. Fusibacter paucivorans gen. nov., sp. nov., an anaerobic, thiosulfate-reducing bacterium from an oil-producing well. Int J Syst Bacteriol. 1999;49:1141–1147. doi: 10.1099/00207713-49-3-1141. [DOI] [PubMed] [Google Scholar]
- Liaw HJ, Mah RA. Isolation and Characterization of Haloanaerobacter chitinovorans gen. nov., sp. nov., a Halophilic, Anaerobic, Chitinolytic Bacterium from a Solar Saltern. Appl Environ Microbiol. 1992;58:260–266. doi: 10.1128/aem.58.1.260-266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhupathiraju VK, McInerney MJ, Woese CR, Tanner RS. Haloanaerobium kushneri sp. nov., an obligately halophilic, anaerobic bacterium from an oil brine. Int J Syst Bacteriol. 1999;49(3):953–960. doi: 10.1099/00207713-49-3-953. [DOI] [PubMed] [Google Scholar]
- Kivistoe A, Santala V, Karp M. Hydrogen production from glycerol using halophilic fermentative bacteria. Bioresource Technol. 2010;101:8671–8677. doi: 10.1016/j.biortech.2010.06.066. [DOI] [PubMed] [Google Scholar]
- Tsai CR, Garcia JL, Patel BK, Cayol JL, Baresi L, Mah RA. Haloanaerobium alcaliphilum sp. nov., an anaerobic moderate halophile from the sediments of Great Salt Lake, Utah. Int J Syst Bacteriol. 1995;45:301–307. doi: 10.1099/00207713-45-2-301. [DOI] [PubMed] [Google Scholar]
- Minnan L, Jinli H, Xiaobin W, Huijuan X, Jinzao C, Chuannan L, Fengzhang Z, Liangshu X. Isolation and characterization of a high H2-producing strain Klebsiella oxytoca HP1 from a hot spring. Res Microbiol. 2005;156:76–81. doi: 10.1016/j.resmic.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Steuber J, Krebs W, Bott M, Dimroth P. A membrane-bound NAD(P) + −reducing hydrogenase provides reduced pyridine nucleotides during citrate fermentation by Klebsiella pneumoniae. J Bacteriol. 1999;181:241–245. doi: 10.1128/jb.181.1.241-245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebl H, Schwab-Hanisch H, Sproer C, Lunsdorf H. Propionispora vibrioides, nov. gen., nov. sp., a new gram-negative, spore-forming anaerobe that ferments sugar alcohols. Arch Microbiol. 2000;174:239–247. doi: 10.1007/s002030000198. [DOI] [PubMed] [Google Scholar]
- Wolin MJ, Miller TL. Molybdate and sulfide inhibit H2 and increase formate production from glucose by Ruminococcus albus. Arch Microbiol. 1980;124:137–142. doi: 10.1007/BF00427718. [DOI] [PubMed] [Google Scholar]
- Mountfort DO, Kaspar HF. Palladium-mediated hydrogenation of unsaturated hydrocarbons with hydrogen gas released during anaerobic cellulose degradation. Appl Environ Microb. 1986;52:744–750. doi: 10.1128/aem.52.4.744-750.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulou G, Ntaikou I, Gavala HN, Skiadas IV, Angelopoulos K, Lyberatos G. Biohydrogen production from sweet sorghum biomass using mixed acidogenic cultures and pure cultures of Ruminococcus albus. Global NEST J. 2007;9:144–151. [Google Scholar]
- Ntaikou I, Gavala HN, Kornaros M, Lyberatos G. Hydrogen production from sugars and sweet sorghum biomass using Ruminococcus albus. Int J Hydrogen Energ. 2008;33:1153–1163. [Google Scholar]
- Ntaikou I, Koutros E, Kornaros M. Valorisation of wastepaper using the fibrolytic/hydrogen producing bacterium Ruminococcus albus. Bioresource Technol. 2009;100:5928–5933. doi: 10.1016/j.biortech.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Latham MJ, Wolin MJ. Fermentation of cellulose by Ruminococcus flavefaciens in the presence and absence of Methanobacterium ruminantium. Appl Environ Microb. 1977;34:297–301. doi: 10.1128/aem.34.3.297-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wolin MJ. Influence of CH4 production by Methanobacterium ruminantium on the fermentation of glucose and lactate by Selenomonas ruminantium. Appl Environ Microbiol. 1977;34:756–759. doi: 10.1128/aem.34.6.756-759.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PH, Morgan HW. Glucose catabolism by Spirochaeta thermophila RI 19..B1. J Bacteriol. 1992;174:2449–2453. doi: 10.1128/jb.174.8.2449-2453.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Song L, Dong X. Sporacetigenium mesophilum gen. nov., sp. nov., isolated from an anaerobic digester treating municipal solid waste and sewage. Int J Syst Evol Microbiol. 2006;56:721–725. doi: 10.1099/ijs.0.63686-0. [DOI] [PubMed] [Google Scholar]
- Hao X, Ma K. Minimal sulfur requirement for growth and sulfur-dependent metabolism of the hyperthermophilic archaeon Staphylothermus marinus. Archaea. 2003;1:191–197. doi: 10.1155/2003/626017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossner AS, Devereux R, Ohnemuller N, Acker G, Stackebrandt E, Drake HL. Thermicanus aegyptius gen. nov., sp. nov., isolated from oxic soil, a fermentative microaerophile that grows commensally with the thermophilic acetogen Moorella thermoacetica. Appl Environ Microb. 1999;65:5124–5133. doi: 10.1128/aem.65.11.5124-5133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J, Ljungdahl LG. Thermoanaerobacter ethanolicus gen. nov; spec. nov; a new, extreme thermophilic, anaerobic bacterium. Arch Microbiol. 1981;128:343–348. doi: 10.1007/BF00405910. [DOI] [Google Scholar]
- Fardeau ML, Faudon C, Cayol JL, Magot M, Patel BKC, Ollivier B. Effect of thiosulfate as electron acceptor on glucose and xylose oxidation by Thermoanaerobacter finnii and a Thermoanaerobacter sp. isolated from oil field water. Res Microbiol. 1996;147:159–165. doi: 10.1016/0923-2508(96)80215-4. [DOI] [PubMed] [Google Scholar]
- Larsen L, Nielsen P, Ahring BK. Thermoanaerobacter mathranii sp. nov., an ethanol-producing, extremely thermophilic anaerobic bacterium from a hot spring in Iceland. Arch Microbiol. 1997;168:114–119. doi: 10.1007/s002030050476. [DOI] [PubMed] [Google Scholar]
- Xue Y, Xu Y, Liu Y, Ma Y, Zhou P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int J Syst Evol Microbiol. 2001;51:1335–1341. doi: 10.1099/00207713-51-4-1335. [DOI] [PubMed] [Google Scholar]
- O-Thong S, Prasertsan P, Karakashev D, Angelidaki I. Thermophilic fermentative hydrogen production by the newly isolated Thermoanaerobacterium thermosaccharolyticum PSU-2. Int J Hydrogen Energ. 2008;33:1204–1214. doi: 10.1016/j.ijhydene.2007.12.015. [DOI] [Google Scholar]
- Ren N, Cao G, Wang A, Lee D-J, Guo W, Zhu Y. Dark fermentation of xylose and glucose mix using isolated Thermoanaerobacterium thermosaccharolyticum W16. Int J Hydrogen Energ. 2008;33:6124–6132. doi: 10.1016/j.ijhydene.2008.07.107. [DOI] [Google Scholar]
- Zavarzina DG, Zhilina TN, Tourova TP, Kuznetsov BB, Kostrikina NA, Bonch-Osmolovskaya EA. Thermanaerovibrio velox sp. nov., a new anaerobic, thermophilic, organotrophic bacterium that reduces elemental sulfur, and emended description of the genus Thermanaerovibrio. Int J Syst Evol Microbiol. 2000;50:1287–1295. doi: 10.1099/00207713-50-3-1287. [DOI] [PubMed] [Google Scholar]
- Eriksen NT, Riis ML, Holm NK, Iversen N. H2 synthesis from pentoses and biomass in Thermotoga spp. Biotechnol Lett. 2011;33:293–300. doi: 10.1007/s10529-010-0439-x. [DOI] [PubMed] [Google Scholar]
- Schröder C, Selig M, Schonheit P. Glucose fermentation to acetate, CO2 and H2 in the anaerobic hyperthermophilic eubacterium Thermotaga maritima: involvement of the Embden-Meyerhof pathway. Arch Microbiol. 1994;161:460–470. [Google Scholar]
- Takahata Y, Nishijima M, Hoaki T, Maruyama T. Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int J Syst Evol Microbiol. 2001;51:1901–1909. doi: 10.1099/00207713-51-5-1901. [DOI] [PubMed] [Google Scholar]
- Munro SA, Zinder SH, Walker LP. The fermentation stoichiometry of Thermotoga neapolitana and influence of temperature, oxygen, and pH on hydrogen production. Biotechnol Prog. 2009;25:1035–1042. doi: 10.1002/btpr.201. [DOI] [PubMed] [Google Scholar]
- Eriksen NT, Nielsen TM, Iversen N. Hydrogen production in anaerobic and microaerobic Thermotoga neapolitana. Biotechnol Lett. 2008;30:103–109. doi: 10.1007/s10529-007-9520-5. [DOI] [PubMed] [Google Scholar]
- Nguyen T-AD, Han S-J, Kim J-P, Kim M-S, Sim S-J. Hydrogen production of the hyperthermophilic eubacterium, Thermotoga neapolitana under N2 sparging condition. Bioresource Technol. 2009;101:S38–S41. doi: 10.1016/j.biortech.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Van Ooteghem Suellen A, Beer Stephen K, Yue Paul C. Hydrogen production by the thermophilic bacterium Thermotoga neapolitana. Appl Biochem Biotechnol. 2002;98-100:177–189. doi: 10.1385/abab:98-100:1-9:177. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Plugge CM, Akkermans ADL, de Vos WM. Victivallis vadensis gen. nov., sp. nov., a sugar-fermenting anaerobe from human faeces. Int J Syst Evol Microbiol. 2003;53:211–215. doi: 10.1099/ijs.0.02362-0. [DOI] [PubMed] [Google Scholar]
- Klibanov AM, Alberti BN, Zale SE. Enzymic synthesis of formic acid from hydrogen and carbon dioxide and production of hydrogen from formic acid. Biotechnol Bioeng. 1982;24:25–36. doi: 10.1002/bit.260240104. [DOI] [PubMed] [Google Scholar]
- Porwal S, Kumar T, Lal S, Rani A, Kumar S, Cheema S, Purohit HJ, Sharma R, Patel SKS, Kalia VC. Hydrogen and polyhydroxybutyrate producing abilities of microbes from diverse habitats by dark fermentative process. Bioresource Technol. 2008;99:5444–5451. doi: 10.1016/j.biortech.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Kotay SM, Das D. Microbial hydrogen production with Bacillus coagulans IIT-BT S1 isolated from anaerobic sewage sludge. Bioresource Technol. 2007;98:1183–1190. doi: 10.1016/j.biortech.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Manikkandan TR, Dhanasekar R, Thirumavalavan K. Microbial production of Hydrogen from sugarcane Bagasse using Bacillus Sp. Int J ChemTech Res. 2009;1:344–348. [Google Scholar]
- Chadwick LJ, Irgens RL. Hydrogen gas production by an Ectothiorhodospira vacuolata strain. Appl Environ Microb. 1991;57:594–596. doi: 10.1128/aem.57.2.594-596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrije T, Bakker RR, Budde MAW, Lai MH, Mars AE, Claassen PAM. Efficient hydrogen production from the lignocellulosic energy crop Miscanthus by the extreme thermophilic bacteria Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Biotechnol Biofuels. 2009;2:12. doi: 10.1186/1754-6834-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotopoulos IA, Bakker RR, de Vrije T, Koukios EG, Claassen PAM. Pretreatment of sweet sorghum bagasse for hydrogen production by Caldicellulosiruptor saccharolyticus. Int J Hydrogen Energ. 2010;35:7738–7747. doi: 10.1016/j.ijhydene.2010.05.075. [DOI] [Google Scholar]
- Mars AE, Veuskens T, Budde MAW, van Doeveren PFNM, Lips SJ, Bakker RR, de Vrije T, Claassen PAM. Biohydrogen production from untreated and hydrolyzed potato steam peels by the extreme thermophiles Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Int J Hydrogen Energ. 2010;35:7730–7737. doi: 10.1016/j.ijhydene.2010.05.063. [DOI] [Google Scholar]
- Alshiyab H, Kalil MS, Hamid AA, Yusoff WMW. Effect of some environmental parameters on hydrogen production using C. acetobutylicum. Pak J Biol Sci. 2008;11:2073–2082. doi: 10.3923/pjbs.2008.2073.2082. [DOI] [PubMed] [Google Scholar]
- Alshiyab H, Kalil MS, Hamid AA, Yusoff WMW. Effect of salts addition on hydrogen production by C. acetobutylicum. Pak J Biol Sci. 2008;11:2193–2200. doi: 10.3923/pjbs.2008.2193.2200. [DOI] [PubMed] [Google Scholar]
- Taguchi F, Chang JD, Mizukami N, Saito-Taki T, Hasegawa K, Morimoto M. Isolation of a hydrogen-producing bacterium, Clostridium beijerinckii strain AM21B, from termites. Can J Microbiol. 1993;39:726–730. doi: 10.1139/m93-105. [DOI] [Google Scholar]
- Cai G, Jin B, Saint C, Monis P. Genetic manipulation of butyrate formation pathways in Clostridium butyricum. J Biotechnol. 2011;155:269–274. doi: 10.1016/j.jbiotec.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Wang X, Jin B, Mulcahy D. Impact of carbon and nitrogen sources on hydrogen production by a newly isolated Clostridium butyricum W5. Int J Hydrogen Energ. 2008;33:4998–5005. doi: 10.1016/j.ijhydene.2008.07.078. [DOI] [Google Scholar]
- Wang X, Jin B. Process optimization of biological hydrogen production from molasses by a newly isolated Clostridium butyricum W5. J Biosci Bioeng. 2009;107:138–144. doi: 10.1016/j.jbiosc.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Evvyernie D, Yamazaki S, Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Identification and characterization of Clostridium paraputrificum M-21, a chitinolytic, mesophilic and hydrogen-producing bacterium. J Biosci Bioeng. 2000;89:596–601. doi: 10.1016/S1389-1723(00)80063-8. [DOI] [PubMed] [Google Scholar]
- Evvyernie D, Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Conversion of chitinous wastes to hydrogen gas by Clostridium paraputrificum M-21. J Biosci Bioeng. 2001;91:339–343. doi: 10.1263/jbb.91.339. [DOI] [PubMed] [Google Scholar]
- Koskinen PEP, Beck SR, Orlygsson J, Puhakka JA. Ethanol and hydrogen production by two thermophilic, anaerobic bacteria isolated from Icelandic geothermal areas. Biotechnol Bioeng. 2008;101:679–690. doi: 10.1002/bit.21942. [DOI] [PubMed] [Google Scholar]
- Li Y-F, Ren N-Q, Yang C-P, Wang A-J, Zadsar M, Li J-Z, Hu L-J. Molecular Characterization and Hydrogen Production of a New Species of Anaerobe. J Environ Sci Health Part A Toxic/Hazard Subst Environ Eng. 2005;40:1929–1938. doi: 10.1080/10934520500184483. [DOI] [PubMed] [Google Scholar]
- Wang X, Hoefel D, Saint CP, Monis PT, Jin B. The isolation and microbial community analysis of hydrogen producing bacteria from activated sludge. J Appl Microbiol. 2007;103:1415–1423. doi: 10.1111/j.1365-2672.2007.03370.x. [DOI] [PubMed] [Google Scholar]
- Jo JH, Lee DS, Park JM. The effects of pH on carbon material and energy balances in hydrogen-producing Clostridium tyrobutyricum JM1. Bioresource Technol. 2008;99:8485–8491. doi: 10.1016/j.biortech.2008.03.060. [DOI] [PubMed] [Google Scholar]
- Jo JH, Lee DS, Kim J, Park JM. Effect of initial glucose concentrations on carbon and energy balances in hydrogen producing Clostridium tyrobutyricum JM1. J Microbiol Biotechnol. 2009;19:291–298. [PubMed] [Google Scholar]
- Sakai S, Yagishita T. Microbial production of hydrogen and ethanol from glycerol-containing wastes discharged from a biodiesel fuel production plant in a bioelectrochemical reactor with thionine. Biotechnol Bioeng. 2007;98:340–348. doi: 10.1002/bit.21427. [DOI] [PubMed] [Google Scholar]
- Kurokawa T, Tanisho S. Effects of Formate on Fermentative Hydrogen Production by Enterobacter aerogenes. Mar Biotechnol. 2005;7:112–118. doi: 10.1007/s10126-004-3088-z. [DOI] [PubMed] [Google Scholar]
- Tanisho S, Suzuki Y, Wakao N. Fermentative hydrogen evolution by Enterobacter aerogenes strain E..82005. Int J Hydrogen Energ. 1987;12:623–627. doi: 10.1016/0360-3199(87)90003-6. [DOI] [Google Scholar]
- Tanisho S, Kuromoto M, Kadokura N. Effect of CO2 removal on hydrogen production by fermentation. Int J Hydrogen Energ. 1998;23:559–563. doi: 10.1016/S0360-3199(97)00117-1. [DOI] [Google Scholar]
- Ren Y, Wang J, Liu Z, Ren Y, Li G. Hydrogen production from the monomeric sugars hydrolyzed from hemicellulose by Enterobacter aerogenes. Renew Energ. 2009;34:2774–2779. doi: 10.1016/j.renene.2009.04.011. [DOI] [Google Scholar]
- Fabiano B, Perego P. Thermodynamic study and optimization of hydrogen production by Enterobacter aerogenes. Int J Hydrogen Energ. 2001;27:149–156. [Google Scholar]
- Converti A, Perego P. Use of carbon and energy balances in the study of the anaerobic metabolism of Enterobacter aerogenes at variable starting glucose concentrations. Appl Microbiol Biot. 2002;59:303–309. doi: 10.1007/s00253-002-1009-5. [DOI] [PubMed] [Google Scholar]
- Perego P, Fabiano B, Ponzano GP, Palazzi E. Experimental study of hydrogen kinetics from agroindustrial byproduct: Optimal conditions for production and fuel cell feeding. Bioprocess Eng. 1998;19:205–211. doi: 10.1007/s004490050507. [DOI] [Google Scholar]
- Shin J-H, Yoon J-H, Lee S-H, Park T-H. Hydrogen production from formic acid in pH-stat fed-batch operation for direct supply to fuel cell. Bioresource Technol. 2009;101:S53–S58. doi: 10.1016/j.biortech.2009.03.032. [DOI] [PubMed] [Google Scholar]
- Shin J-H, Yoon JH, Ahn EK, Kim M-S, Sim SJ, Park TH. Fermentative hydrogen production by the newly isolated Enterobacter asburiae SNU-1. Int J Hydrogen Energ. 2007;32:192–199. doi: 10.1016/j.ijhydene.2006.08.013. [DOI] [Google Scholar]
- Kumar N, Ghosh A, Das D. Redirection of biochemical pathways for the enhancement of H2 production by Enterobacter cloacae. Biotechnol Lett. 2001;23:537–541. doi: 10.1023/A:1010334803961. [DOI] [Google Scholar]
- Nath K, Kumar A, Das D. Hydrogen production by Rhodobacter sphaeroides strain O..U..001 using spent media of Enterobacter cloacae strain DM11. Appl Microbiol Biot. 2005;68:533–541. doi: 10.1007/s00253-005-1887-4. [DOI] [PubMed] [Google Scholar]
- Nath K, Kumar A, Das D. Effect of some environmental parameters on fermentative hydrogen production by Enterobacter cloacae DM11. Can J Microbiol. 2006;52:525–532. doi: 10.1139/w06-005. [DOI] [PubMed] [Google Scholar]
- Nath K, Muthukumar M, Kumar A, Das D. Kinetics of two-stage fermentation process for the production of hydrogen. Int J Hydrogen Energ. 2008;33:1195–1203. doi: 10.1016/j.ijhydene.2007.12.011. [DOI] [Google Scholar]
- Kumar N, Das D. Enhancement of hydrogen production by Enterobacter cloacae IIT-BT 08. Process Biochem. 2000;35:589–593. doi: 10.1016/S0032-9592(99)00109-0. [DOI] [Google Scholar]
- Higgins TE, Johnson MJ. Pathways of anaerobic acetate utilization in Escherichia coli and Aerobacter cloacae. J Bacteriol. 1970;101:885–891. doi: 10.1128/jb.101.3.885-891.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood AC, Ledingham GA, Neish AC. Dissimilation of glucose at controlled pH values by pigmented and non-pigmented strains of Escherichia coli. J Bacteriol. 1956;72:497–499. doi: 10.1128/jb.72.4.497-499.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. Efficient induction of formate hydrogen lyase of aerobically grown Escherichia coli in a three-step biohydrogen production process. Appl Microbiol Biot. 2007;74:754–760. doi: 10.1007/s00253-006-0721-y. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. Enhanced hydrogen production from glucose using ldh- and frd-inactivated Escherichia coli strains. Appl Microbiol Biot. 2006;73:67–72. doi: 10.1007/s00253-006-0456-9. [DOI] [PubMed] [Google Scholar]
- Chaudhary N, Ngadi MO, Simpson BK, Kassama LS. Biosynthesis of ethanol and hydrogen by glycerol fermentation using Escherichia coli. Adv Chem Eng Sci. 2011;1:83–89. doi: 10.4236/aces.2011.13014. [DOI] [Google Scholar]
- Chittibabu G, Nath K, Das D. Feasibility studies on the fermentative hydrogen production by recombinant Escherichia coli BL-21. Process Biochem. 2006;41:682–688. doi: 10.1016/j.procbio.2005.08.020. [DOI] [Google Scholar]
- Penfold DW, Macaskie LE. Production of H2 from sucrose by Escherichia coli strains carrying the pUR400 plasmid, which encodes invertase activity. Biotechnol Lett. 2004;26:1879–1883. doi: 10.1007/s10529-004-6035-1. [DOI] [PubMed] [Google Scholar]
- Redwood MD, Macaskie LE. A two-stage, two-organism process for biohydrogen from glucose. Int J Hydrogen Energ. 2006;31:1514–1521. doi: 10.1016/j.ijhydene.2006.06.018. [DOI] [Google Scholar]
- Orozco RL, Redwood MD, Yong P, Caldelari I, Sargent F, Macaskie LE. Towards an integrated system for bio-energy: Hydrogen production by Escherichia coli and use of palladium-coated waste cells for electricity generation in a fuel cell. Biotechnol Lett. 2011;32:1837–1845. doi: 10.1007/s10529-010-0383-9. [DOI] [PubMed] [Google Scholar]
- Murarka A, Dharmadi Y, Yazdani SS, Gonzalez R. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl Environ Microb. 2008;74:1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala G, Stetter KO. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 DegC. Arch Microbiol. 1986;145:56–61. doi: 10.1007/BF00413027. [DOI] [Google Scholar]
- Cao G, Ren N, Wang A, Lee D-J, Guo W, Liu B, Feng Y, Zhao Q. Acid hydrolysis of corn stover for biohydrogen production using Thermoanaerobacterium thermosaccharolyticum W16. Int J Hydrogen Energ. 2009;34:7182–7188. doi: 10.1016/j.ijhydene.2009.07.009. [DOI] [Google Scholar]
- Ueno Y, Haruta S, Ishii M, Igarashi Y. Characterization of a microorganism isolated from the effluent of hydrogen fermentation by microflora. J Biosci Bioeng. 2001;92:397–400. doi: 10.1263/jbb.92.397. [DOI] [PubMed] [Google Scholar]
- Teplyakov VV, Gassanova LG, Sostina EG, Slepova EV, Modigell M, Netrusov AI. Lab-scale bioreactor integrated with active membrane system for hydrogen production: experience and prospects. Int J Hydrogen Energ. 2002;27:1149–1155. doi: 10.1016/S0360-3199(02)00093-9. [DOI] [Google Scholar]
- Huber R, Langworthy TA, Koenig H, Thomm M, Woese CR, Sleytr UB, Stetter KO. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 DegC. Arch Microbiol. 1986;144:324–333. doi: 10.1007/BF00409880. [DOI] [Google Scholar]
- Ren N, Wang A, Gao L, Xin L, Lee D-J, Su A. Bioaugmented hydrogen production from carboxymethyl cellulose and partially delignified corn stalks using isolated cultures. Int J Hydrogen Energ. 2008;33:5250–5255. doi: 10.1016/j.ijhydene.2008.05.020. [DOI] [Google Scholar]
- Yokoi H, Saitsu A, Uchida H, Hirose J, Hayashi S, Takasaki Y. Microbial hydrogen production from sweet potato starch residue. J Biosci Bioeng. 2001;91:58–63. doi: 10.1263/jbb.91.58. [DOI] [PubMed] [Google Scholar]
- Yokoi H, Maki R, Hirose J, Hayashi S. Microbial production of hydrogen from starch-manufacturing wastes. Biomass Bioenerg. 2002;22:389–395. doi: 10.1016/S0961-9534(02)00014-4. [DOI] [Google Scholar]
- Kamalaskar LB, Dhakephalkar PK, Meher KK, Ranade DR. High biohydrogen yielding Clostridium sp. DMHC-10 isolated from sludge of distillery waste treatment plant. Int J Hydrogen Energ. 2010;35:10639–10644. doi: 10.1016/j.ijhydene.2010.05.020. [DOI] [Google Scholar]
- Zhang H, Bruns MA, Logan BE. Biological hydrogen production by Clostridium acetobutylicum in an unsaturated flow reactor. Water Res. 2006;40:728–734. doi: 10.1016/j.watres.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Vasconcelos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol. 1994;176:1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin H-L, Chen Z-S, Chou CP. Fedbatch Operation Using Clostridium acetobutylicum Suspension Culture as Biocatalyst for Enhancing Hydrogen Production. Biotechnol Prog. 2003;19:383–388. doi: 10.1021/bp0200604. [DOI] [PubMed] [Google Scholar]
- Yokoi H, Maeda Y, Hirose J, Hayashi S, Takasaki Y. H2 production by immobilized cells of Clostridium butyricum on porous glass beads. Biotechnol Tech. 1997;11:431–433. doi: 10.1023/A:1018429109020. [DOI] [Google Scholar]
- Heyndrickx M, De Vos P, De Ley J. Hydrogen production from chemostat fermentation of glucose by Clostridium butyricum and Clostridium pasteurianum in ammonium- and phosphate limitation. Biotechnol Lett. 1990;12:731–736. doi: 10.1007/BF01024730. [DOI] [Google Scholar]
- Heyndrickx M, De Vos P, De Ley J. Fermentation of D-xylose by Clostridium butyricum LMG 1213 t1 in chemostats. Enzyme Microb Technol. 1991;13:893–897. doi: 10.1016/0141-0229(91)90105-J. [DOI] [Google Scholar]
- Saint-Amans S, Girbal L, Andrade J, Ahrens K, Soucaille P. Regulation of carbon and electron flow in Clostridium butyricum VPI 3266 grown on glucose-glycerol mixtures. J Bacteriol. 2001;183:1748–1754. doi: 10.1128/JB.183.5.1748-1754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F, Yamada K, Hasegawa K, Taki-Saito T, Hara K. Continuous hydrogen production by Clostridium sp. strain no. 2 from cellulose hydrolyzate in an aqueous two-phase system. J Ferment Bioeng. 1996;82:80–83. doi: 10.1016/0922-338X(96)89460-8. [DOI] [Google Scholar]
- Taguchi F, Mizukami N, Saito-Taki T, Hasegawa K. Hydrogen production from continuous fermentation of xylose during growth of Clostridium sp. strain No. 2. Can J Microbiol. 1995;41:536–540. doi: 10.1139/m95-071. [DOI] [Google Scholar]
- Collet C, Girbal L, Peringer P, Schwitzguebel J-P, Soucaille P. Metabolism of lactose by Clostridium thermolacticum growing in continuous culture. Arch Microbiol. 2006;185:331–339. doi: 10.1007/s00203-006-0098-4. [DOI] [PubMed] [Google Scholar]
- Collet C, Adler N, Schwitzguebel J-P, Peringer P. Hydrogen production by Clostridium thermolacticum during continuous fermentation of lactose. Int J Hydrogen Energ. 2004;29:1479–1485. doi: 10.1016/j.ijhydene.2004.02.009. [DOI] [Google Scholar]
- Magnusson L, Cicek N, Sparling R, Levin D. Continuous hydrogen production during fermentation of alpha-cellulose by the thermophillic bacterium Clostridium thermocellum. Biotechnol Bioeng. 2009;102:759–766. doi: 10.1002/bit.22092. [DOI] [PubMed] [Google Scholar]
- Vancanneyt M, De Vos P, Vennens L, De Ley J. Lactate and ethanol dehydrogenase activities in continuous cultures of Clostridium thermosaccharolyticum LMG 6564. J Gen Microbiol. 1990;136:1945–1951. doi: 10.1099/00221287-136-10-1945. [DOI] [Google Scholar]
- Jo JH, Lee DS, Park D, Park JM. Biological hydrogen production by immobilized cells of Clostridium tyrobutyricum JM1 isolated from a food waste treatment process. Bioresource Technol. 2008;99:6666–6672. doi: 10.1016/j.biortech.2007.11.067. [DOI] [PubMed] [Google Scholar]
- Mitchell RJ, Kim J-S, Jeon B-S, Sang B-I. Continuous hydrogen and butyric acid fermentation by immobilized Clostridium tyrobutyricum ATCC 25755: Effects of the glucose concentration and hydraulic retention time. Bioresource Technol. 2009;100:5352–5355. doi: 10.1016/j.biortech.2009.05.046. [DOI] [PubMed] [Google Scholar]
- Whang L-M, Lin C-A, Liu IC, Wu C-W, Cheng H-H. Metabolic and energetic aspects of biohydrogen production of Clostridium tyrobutyricum: The effects of hydraulic retention time and peptone addition. Bioresour Technol. 2011;102:8378–8383. doi: 10.1016/j.biortech.2011.03.101. [DOI] [PubMed] [Google Scholar]
- Rachman MA, Nakashimada Y, Kakizono T, Nishio N. Hydrogen production with high yield and high evolution rate by self-flocculated cells of Enterobacter aerogenes in a packed-bed reactor. Appl Microbiol Biot. 1998;49:450–454. doi: 10.1007/s002530051197. [DOI] [Google Scholar]
- Tanisho S, Ishiwata Y. Continuous hydrogen production from molasses by fermentation using urethane foam as a support of flocks. Int J Hydrogen Energ. 1995;20:541–545. doi: 10.1016/0360-3199(94)00101-5. [DOI] [Google Scholar]
- Tanisho S, Ishiwata Y. Continuous hydrogen production from molasses by the bacterium Enterobacter aerogenes. Int J Hydrogen Energ. 1994;19:807–812. doi: 10.1016/0360-3199(94)90197-X. [DOI] [Google Scholar]
- Palazzi E, Fabiano B, Perego P. Process development of continuous hydrogen production by Enterobacter aerogenes in a packed column reactor. Bioprocess Eng. 2000;22:205–213. doi: 10.1007/PL00009112. [DOI] [Google Scholar]
- Palazzi E, Perego P, Fabiano B. Mathematical modelling and optimization of hydrogen continuous production in a fixed bed bioreactor. Chem Eng Sci. 2002;57:3819–3830. doi: 10.1016/S0009-2509(02)00322-6. [DOI] [Google Scholar]
- Turcot J, Bisaillon A, Hallenbeck PC. Hydrogen production by continuous cultures of Escherichia coli under different nutrient regimes. Int J Hydrogen Energ. 2008;33:1465–1470. doi: 10.1016/j.ijhydene.2007.09.034. [DOI] [Google Scholar]
- Solomon BO, Zeng AP, Biebl H, Ejiofor AO, Posten C, Deckwer WD. Effects of substrate limitation on product distribution and H2/CO2 ratio in Klebsiella pneumoniae during anaerobic fermentation of glycerol. Appl Microbiol Biot. 1994;42:222–226. [Google Scholar]
- Chou C-J, Shockley KR, Conners SB, Lewis DL, Comfort DA, Adams MWW, Kelly RM. Impact of substrate glycoside linkage and elemental sulfur on bioenergetics of and hydrogen production by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microb. 2007;73:6842–6853. doi: 10.1128/AEM.00597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicho RN, Ma K, Adams MWW, Kelly RM. Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1993;175:1823–1830. doi: 10.1128/jb.175.6.1823-1830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti EL, Kafkewitz D, Wolin MJ, Bryant MP. Glucose fermentation products in Ruminococcus albus grown in continuous culture with Vibrio succinogenes: changes caused by interspecies transfer of H 2. J Bacteriol. 1973;114:1231–1240. doi: 10.1128/jb.114.3.1231-1240.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Weimer PJ, Ralph J. Formation of formate and hydrogen, and flux of reducing equivalents and carbon in Ruminococcus flavefaciens FD-1. A van Leeuw J Microb. 1997;72:101–109. doi: 10.1023/A:1000256221938. [DOI] [PubMed] [Google Scholar]
- Kanai T, Imanaka H, Nakajima A, Uwamori K, Omori Y, Fukui T, Atomi H, Imanaka T. Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J Biotechnol. 2005;116:271–282. doi: 10.1016/j.jbiotec.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Claassen PAM, Van Lier JB, Contreras AML, Van Niel EWJ, Sijtsma L, Stams AJM, De Vries SS, Weusthuis RA. Utilisation of biomass for the supply of energy carriers. Appl Microbiol Biot. 1999;52:741–755. doi: 10.1007/s002530051586. [DOI] [Google Scholar]
- Noike T, Ko IB, Yokoyama S, Kohno Y, Li YY. Continuous hydrogen production from organic waste. Water Sci Technol. 2005;52:145–151. [PubMed] [Google Scholar]
- Thompson LJ, Gray VM, Kalala B, Lindsay D, Reynolds K, von Holy A. Biohydrogen production by Enterobacter cloacae and Citrobacter freundii in carrier induced granules. Biotechnol Lett. 2008;30:271–274. doi: 10.1007/s10529-007-9527-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains reported to produce biohydrogen without the possibility to calculate or retrieve quantitative results[3,4,6,7,48,80,82,90-204].
Closed batch dark fermentative biohydrogen production[4,29,35,39,42,53-55,64-67,69,78,82,83,85,86,98,120,130,131,137,142,166,180,182-184,192,194,198,205-314].
Batch dark fermentative biohydrogen production[28,32,33,45,49,56,57,62,63,65,66,72,76,78,79,81,84,90,96,110,115,315-369].
Chemostat culture dark fermentative biohydrogen production[29,34,46,47,56,75,77,96,118,122,124,125,228,257-260,279,286,291,292,331,353,356,370-400].
Fed-batch dark fermentative biohydrogen production[38].