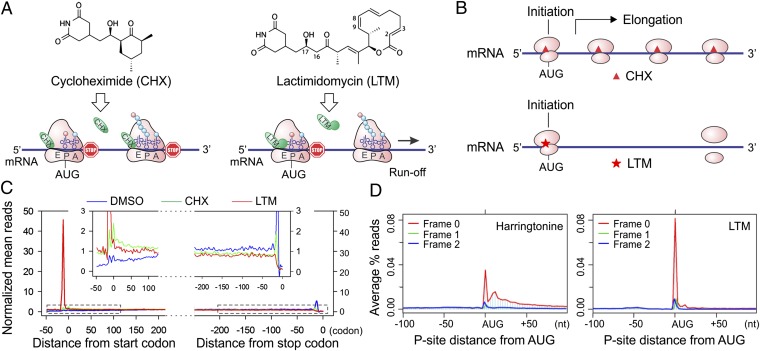
Fig. 1.
Experimental strategy of GTI-seq using ribosome E-site translation inhibitors. (A) Schematic diagram of the experimental design for GTI-seq. Translation inhibitors CHX and LTM bind to the ribosome E-site, resulting in inhibition of translocation. CHX binds to all translating ribosomes (Left), but LTM preferentially incorporates into the initiating ribosomes when the E-site is free of tRNA (Right). (B) Ribosome profiling using CHX and LTM side by side allows the initiating ribosome to be distinguished from the elongating one. (C) HEK293 cells were treated with DMSO, 100 μM CHX, or 50 μM LTM for 30 min before ribosome profiling. Normalized RPF reads are averaged across the entire transcriptome, aligned at either their start site or stop codon from the 5′ end of RPFs. (D) Metagene analysis of RPFs obtained from HEK293 cells treated with harringtonine (Left) or LTM (Right). All mapped reads are aligned at the annotated start codon AUG, and the density of reads at each nucleotide position is averaged using the P-site of RPFs.