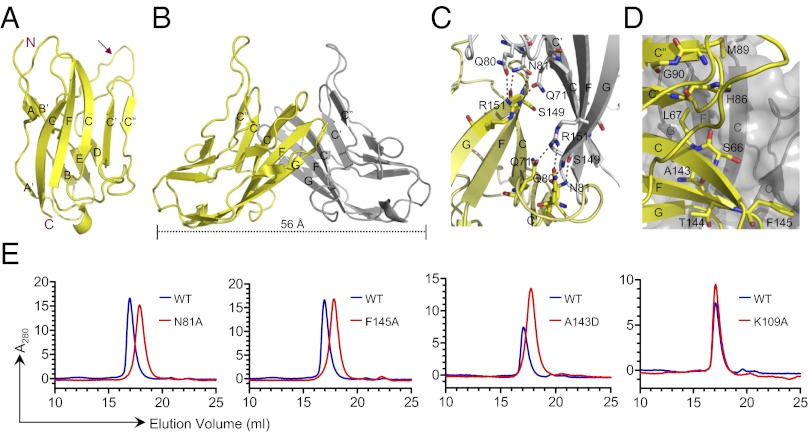
Fig. 2.
(A) Structure of the IgV domain of human nectin-2 showing a classical two-layer β-sandwich topology. The front and back sheets of the domain are composed of the GFCC′C′′ and ABED strands, respectively. The unusual long loop between the D and E strands is identified by an arrow. (B) Two monomers (yellow and gray) interact in a nearly orthogonal fashion to form the dimer, resulting in an end-to-end distance of ∼56 Å. The dimer interface is formed by the orthogonal association of the front β-sheets with predominant contribution from the C, C′, C′′, and F strands of each monomer. (C) Residues participating in hydrogen bonds at the dimer interface are shown in ball-and-stick representation (dashed lines represent hydrogen bonds). (D) Residues participating in van der Waals contacts at the dimer interface are shown by ball-and-stick representation from one molecule only. (E) Confirmation of dimer interface by size-exclusion chromatography. The elution profile of wild-type nectin-2 is shown by the blue lines, and the mutants N81A, F145A, A143D, and K109A are represented by the red lines. In all of the cases except K109A (which is located outside of the dimer interface), the elution volume of the mutants increases significantly.