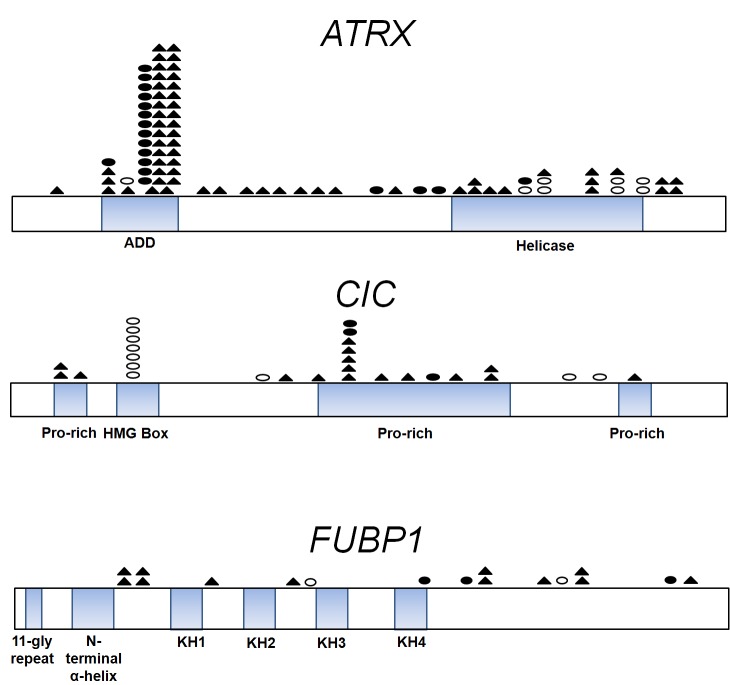
Figure 1. Distribution of ATRX, CIC, and FUBP1 mutations in gliomas.
Schematics of the coding sequences of ATRX, CIC, and FUBP1 are shown. Mutations were predominantly truncating mutations such as frameshifts (shaded triangles) and nonsense mutations (shaded circles). Empty circles correspond to missense mutations. ATRX contains a DNA-binding ATRX-DNMT3-DNMT3L (ADD) domain and an ATPase Helicase Domain. ATRX alterations occurred in 20% of pediatric GBMs (n=25) and all of those mutations clustered in the Helicase domain of ATRX as described previously [14]. ATRX mutations in adult gliomas instead distributed evenly. CIC contains four highly conserved domains, the HMG (high mobility group) box responsible for DNA binding and proline-rich (pro-rich) domains. Among CIC mutations (n=29), a predilection for missense mutations occurred in the HMG box domain (24%). Mutations in FUBP1 (n=16) occurred 3' of the N-terminal α-helix domain, which controls binding of FUBP1 to the FBP-interacting repressor (FIR). Gene representations are to scale.