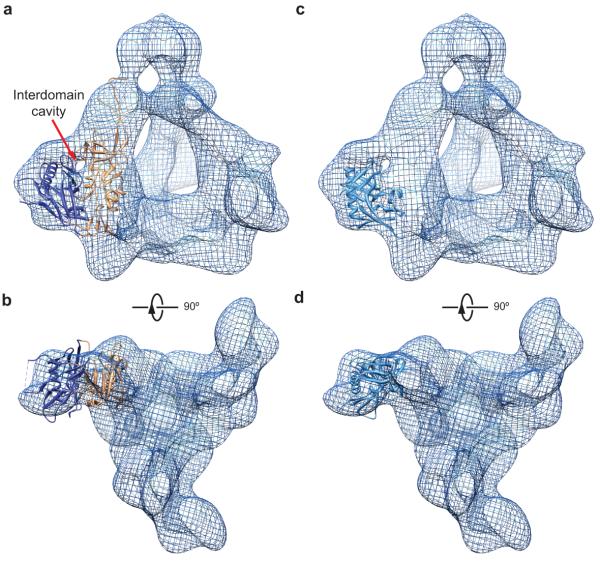
Figure 3.
Fit of the crystal structure of CD4-bound gp120 core into the cryo-EM density of the unliganded Env trimer in two different configurations. (a, b) The CD4-bound gp120 core structure (PDB ID: 3JWD)8 can fit into the local Env trimer cryo-EM density identified by segmentation as the gp120 inner domain and outer domain (See Fig. 2). This fitting configuration represents the best fit of the gp120 core crystal structure, taking into account both the inner and outer domains. The cryo-EM density is shown as a meshwork representation. The red arrow in a marks the interdomain cavity in the gp120 subunit. The gp120 core is missing the V1/V2 and V3 variable regions8. The inner domain and outer domain of the gp120 core crystal structure are coloured orange and dark blue, respectively. A fraction of the residues of the gp120 outer domain does not fit in the cryo-EM density in this fitting configuration, suggesting conformational differences between the unliganded and CD4-bound states. (c, d) The gp120 outer domain from the CD4-bound gp120 core structure8 was fitted into the cryo-EM density of the Env trimer, without consideration of the fit of the gp120 inner domain. This allows a much better fitting of the gp120 outer domain than that seen in a and b; only the LV5 variable loop has a few residues out of the cryo-EM density. Under the same fitting configuration, the inner domain of the CD4-bound gp120 core is largely outside the Env trimer cryo-EM density (not shown). This observation suggests that the gp120 inner domain rotates with respect to the outer domain upon binding CD4.