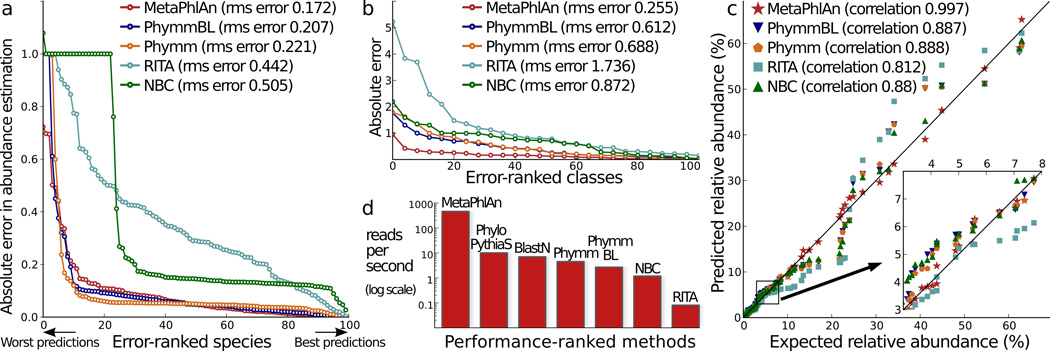
Fig. 1. Comparison of MetaPhlAn to existing methods.
We used 10 total synthetic metagenomes to compare MetaPhlAn to PhymmBL12, BLAST13, theRITA pipeline14, and NBC15. a–b) Absolute and root mean squared (rms) errors with respect to 100 total organisms in one synthetic metagenome at the species (a) and class (b) level c) Correlations of inferred and true species abundances on 8 non-evenly distributed synthetic metagenomes;. d) Reads-per-second rates for the tested methods on single CPUs.