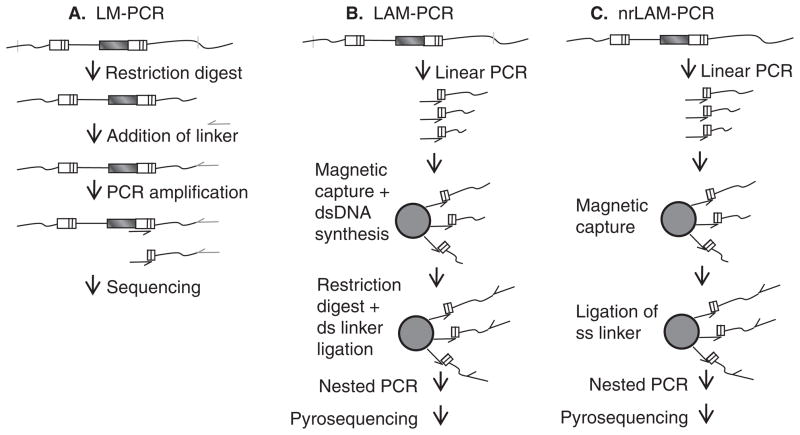
Figure 3. Identification of provirus-chromosome junctions.
A. Ligation-mediated PCR (LM-PCR). Genomic DNA with integrated vector proviruses is digested with frequent-cutting restriction enzymes. A linker is ligated to the cleaved DNA, and after PCR amplification the products are sequenced with a long terminal repeat (LTR)-specific primer. B. Linear-amplification-mediated PCR (LAM-PCR). Linear PCR with only a LTR-specific primer is performed on genomic DNA with integrated vector proviruses to amplify provirus–chromosome junctions. Following amplification, provirus–chromosome junctions of various lengths are produced. The products are enriched on magnetic beads converted to dsDNA, digested with a restriction enzyme (s), typically frequent-cutting enzymes with a 4 bp recognition site, and a double-stranded linker is ligated to the ends. Nested PCR is followed by direct sequencing, shotgun cloning into bacterial plasmids and sequencing or pyrosequencing. C. Non-restrictive linear-amplification-mediated PCR (nrLAM-PCR). Linear PCR with only a LTR-specific primer is performed on genomic DNA with integrated vector proviruses to amplify provirus–chromosome junctions. The products are enriched on magnetic beads and a single-stranded linker is ligated to the ends. Nested PCR is followed by pyrosequencing. For all methods aligning the chromosomal sequence immediately next to the LTR to a published genome allows identification of the provirus integration site.