Abstract
The major polysaccharides in dicot wood biomass are cellulose and xylan. Although wood-associated cellulose synthase genes responsible for cellulose biosynthesis have been characterized, wood-associated xylan synthase genes have not been biochemically identified. A recent report by Lee et al. (2012) provides the first biochemical evidence that two functionally non-redundant Arabidopsis GT43 members are xylosyltransferases (XylTs) that function cooperatively in the elongation of the xylan backbone. We further extend this finding in the current report demonstrating that two poplar (Populus trichocarpa) GT43 glycosyltransferases, PtrGT43B and PtrGT43C, are xylan XylTs involved in wood formation. We show that microsomes from transgenic tobacco BY2 cells coexpressing PtrGT43B and PtrGT43C exhibited a high XylT activity capable of generating β-(1,4)-linked xylooligosaccharides, whereas little XylT activity was detected in microsomes with expression of PtrGT43B or PtrGT43C alone. These findings indicate that poplar GT43 members are XylTs that act cooperatively in catalyzing the successive transfer of xylosyl residues during xylan backbone biosynthesis, which provides further support of the hypothesis that the biochemical functions of GT43 members in vascular plants are evolutionarily conserved.
Keywords: GT43, poplar, wood formation, xylan, xylosyltransferase
Wood, mainly composed of cellulose, hemicelluloses (xylan and mannan) and lignin, is the most abundant biomass and carbon reservoir produced by land plants. Because wood is an abundant raw material widely used in our daily life, there has been an intensive research effort to understand the structure and biosynthesis of wood components in the hope of genetically modifying the quality and quantity of wood to suit our various needs. In particular, much attention has been paid to the study of lignin biosynthesis during wood formation. Transgenic trees with reduced lignin have been shown to have a positive impact on the reduction of chemical usage during pulping.1 Recently, cellulosic biomass from trees is considered to be an abundant source for biofuel production.2 Because polysaccharides from wood biomass are the source of sugars used for fermentation into biofuels, it is critical to have a complete understanding of the biosynthesis of cellulose and hemicelluloses in addition to lignin in order to develop strategies to custom-design wood cell wall composition tailored for biofuel production.3 Cellulose synthase genes responsible for wood cellulose biosynthesis have been functionally characterized in poplar.4 Wood-associated mannan synthase genes involved in the biosynthesis of mannan, a minor hemicellulose component in dicot wood but a major one in gymnosperm wood,5 have also been biochemically analyzed.6,7 Recent molecular and genetic studies of wood-associated glycosyltransferase genes have provided a first glimpse into the biosynthesis of xylan, the major hemicellulose in dicot wood.
Xylan from dicot wood is made of a linear β-(1,4)-linked xylosyl residues with a degree of polymerization of up to 120.8 About 20% of the xylosyl residues on the xylan backbone are substituted at O-2 with methylglucuronic acid and are often heavily acetylated at C-2 and C-3.5 The reducing end of xylan from both dicot wood and gymnosperm wood has been shown to contain a unique tetrasaccharide sequence β-d-Xylp-(1→3)-α-l-Rhap-(1→2)-α-d-GalpA-(1→4)-d-Xylp.9-13 Based on the xylan structure, it is estimated that at least 8 enzymes are needed for the biosynthesis of the reducing end tetrasaccharide sequence, the elongation of the xylan backbone, and the addition and modification of side chains. Previous transcriptome profiling of developing wood in poplar revealed that the expression of 25 glycosyltransferase genes is associated with secondary wall biosynthesis.14 Among them, GT43 members have been proven to be functional orthologs of Arabidopsis IRX9/IRX1412,15-18 involved in the elongation of xylan backbone,19,20 and GT47C, GT8D and GT8E/F are functional orthologs of Arabidopsis FRA8,21 IRX812,15 and PARVUS,15,22 respectively, which are essential for the biosynthesis of the xylan reducing end sequence.13,23,24 However, biochemical proof of the enzymatic activities of these wood-associated glycosyltransferases is still lacking and the identity of xylan synthases responsible for xylan backbone biosynthesis remains to be identified.
It was previously proposed that cellulose synthase-like genes (Csls) are strong candidates for xylan synthases,25,26 but none of the Csl genes except the mannan synthase genes CslAs has been found to be induced during wood formation in poplar14 and thus Csls are unlikely xylan synthase genes. Recently, Lee et al.27 reported that two Arabidopsis GT43 members, IRX9 and IRX14, are XylTs that act together in successive transfer of xylosyl residues onto the xylooligomer acceptors, which provides biochemical evidence demonstrating that GT43 members are part of the long-sought xylan synthase complex responsible for the xylan backbone elongation. Family GT43 glycosyltransferase genes are present in the genomes of all vascular plants thus far sequenced and it was hypothesized that the biochemical functions of GT43 members in vascular plants are conserved.27 To test this hypothesis, we preformed enzymatic studies of two functionally non-redundant poplar GT43 members, PtrGT43B and PtrGT43C, by heterologously expressing them in tobacco BY cells and demonstrated that microsomes from transgenic BY2 cells coexpressing PtrGT43B and PtrGT43C are capable of transferring multiple β-(1,4)-linked xylosyl residues onto xylooligomer acceptors.
We first analyzed the XylT activity of microsomes from the developing wood of poplar. It was demonstrated that microsomes isolated from poplar stems undergoing active wood formation efficiently transferred 14C-labeled xylosyl residues onto the xylotetraose (Xyl4) acceptor in a concentration-dependent manner (Fig. 1A). The XylT activity was found to be both time- and protein concentration-dependent (Fig. 1B and C). The apparent Km and Vmax of the XylT activity were 5.9 mM and 18.1 pmol/min/mg protein, respectively. Using fluorescent anthranilic acid (AA)-labeled xylooligomers as acceptors, we revealed that although the XylT activity could not use the xylose monomer as an acceptor, it readily transferred xylosyl residues onto xylooligomers ranging from 2 to 6 residues, resulting in the production of xylooligosaccharides with up to 11 xylosyl residues after a 30 min incubation (Fig. 2). These data demonstrate that microsomes from poplar developing wood possess a xylan XylT activity that is capable of transferring multiple xylosyl residues onto xylooligomer acceptors.
Figure 1.
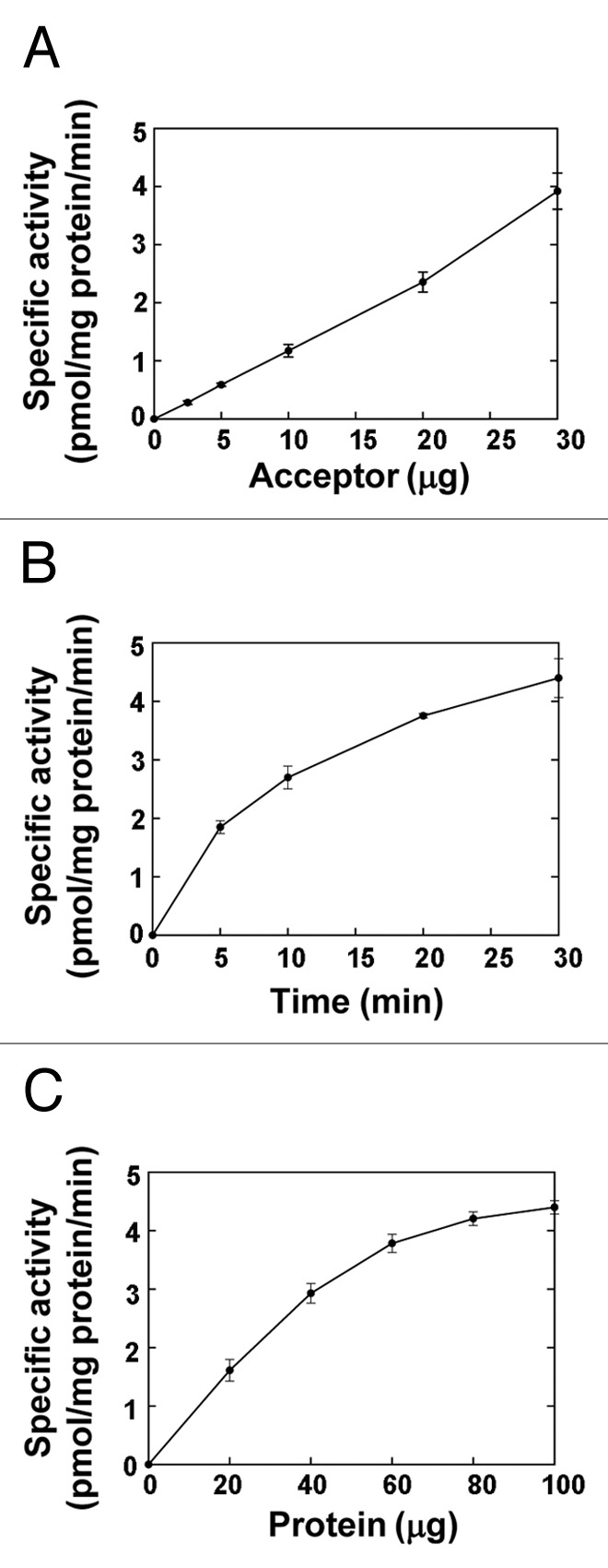
Biochemical properties of xylan XylT activity in microsomes from poplar stems. The XylT activity was measured by incubating microsomes with UDP-[14C]-Xyl and Xyl4 and counting the amount of radioactivity of the reaction products according to Lee et al.16 All assays were repeated twice and the data are means ± SE. (A) Dependence of the XylT activity on the acceptor concentration. (B) Time course of the transfer of the radiolabeled xylosyl residues onto the acceptor by the XylT activity. (C) The XylT activity is protein concentration-dependent.
Figure 2.
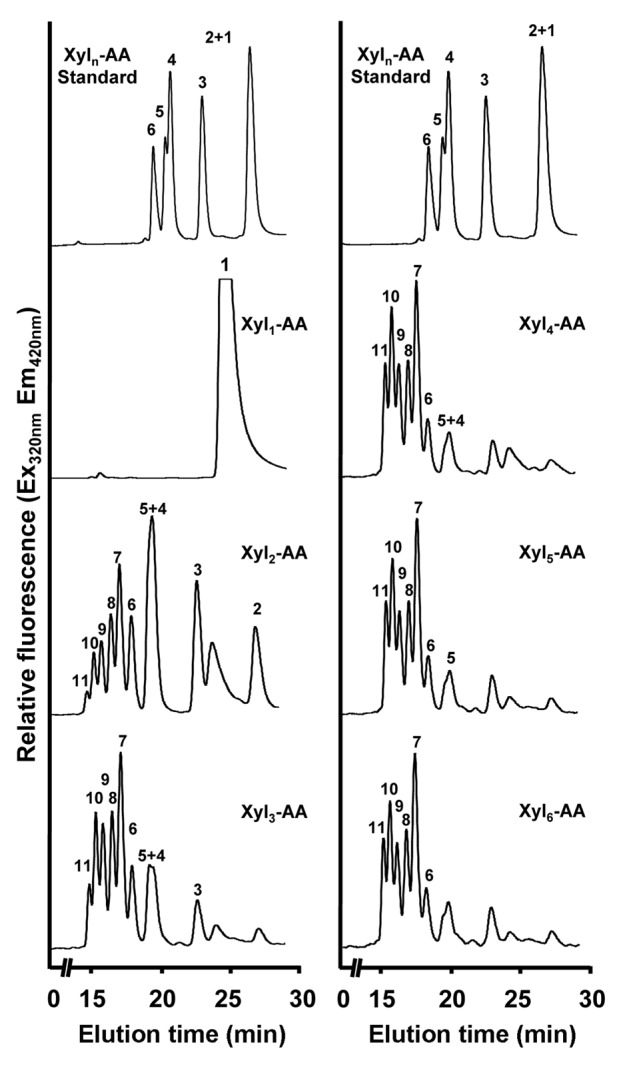
Xylooligomers of various lengths as acceptors for the XylT activity of poplar stems. The XylT activity was assayed by incubating the microsomes from poplar stems with UDP-Xyl and the acceptor Xyl1-AA, Xyl2-AA, Xyl3-AA, Xyl4-AA, Xyl5-AA, or Xyl6-AA, and the reaction products were analyzed by reverse-phase HPLC and detected for fluorescent signals according to Lee et al.16 A chromatogram of standard Xyl1–6-AA is shown at the top.
Poplar GT43 members have previously been shown to form two functionally non-redundant groups that are proposed to be involved in xylan backbone biosynthesis.20 To investigate the enzymatic activity of poplar GT43 members, we chose one representative, PtrGT43B and PtrGT43C, from each of the two functionally non-redundant GT43 groups and heterologously expressed them in tobacco BY2 cells for XylT activity assay. We created four groups of transgenic cell lines including PtrGT43B overexpressors, PtrGT43C overexpressors, PtrGT43B and PtrGT43C co-overexpressors, and the control transformed with an empty vector. For each group, more than 60 independent transgenic lines were generated. Microsomes isolated from these transgenic lines were used for XylT activity assay with Xyl4 as an acceptor and 14C-UDP-Xylose as a donor and the representative data of 10 cell lines from each group were shown in Figure 3A. It was found that while microsomes with expression of PtrGT43B or PtrGT43C alone did not exhibit any difference in the XylT activity compared with those of the control, coexpression of PtrGT43B and PtrGT43C resulted in a significant increase in the incorporation of 14C-xylose onto Xyl4 (Fig. 3A). In particular, two lines of PtrGT43B/C coexpressors displayed a XylT activity equivalent to about 70% of that of microsomes from poplar stems. The XylT activity of microsomes from the PtrGT43B and PtrGT43C coexpressors was dependent of the Xyl4 acceptor concentration (Fig. 3B) with an apparent Km of 5.27 mM and Vmax of 7.3 pmol/min/mg protein, which are similar to the values of Km and Vmax exhibited by the XylT activity of microsomes from poplar stems. These results established that coexpression of PtrGT43B and PtrGT43C reconstitutes a XylT activity that is capable of transferring xylosyl residues onto xylooligomer acceptors.
Figure 3.
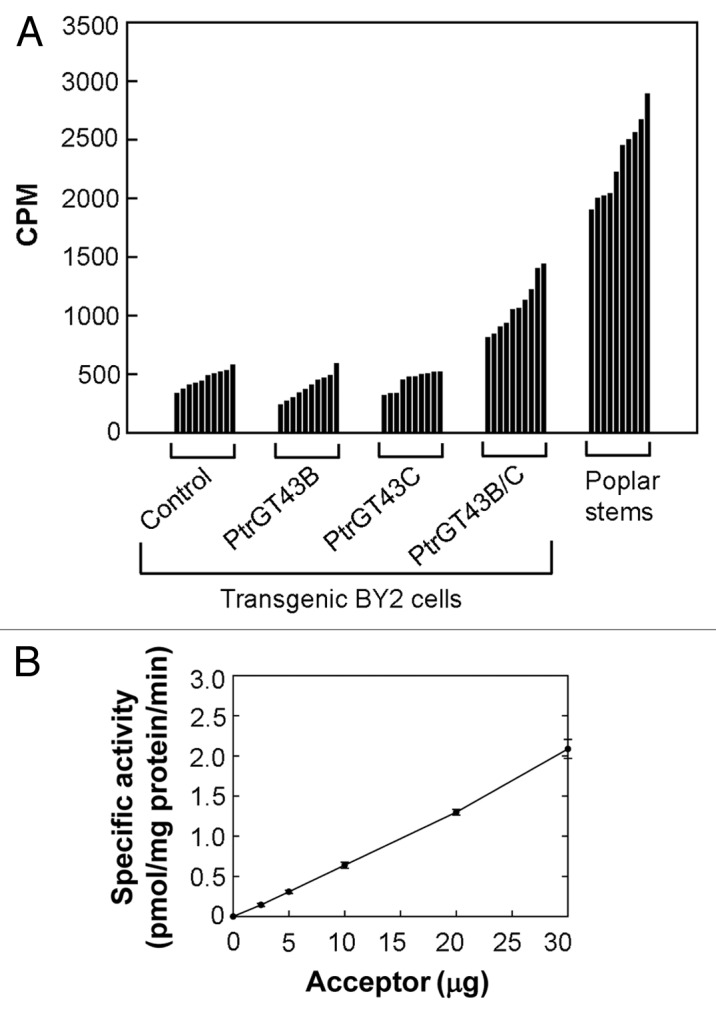
Detection of xylan XylT activity in transgenic BY2 cells expressing PtrGT43B and PtrGT43C. The XylT activity was measured by incubating microsomes with UDP-[14C]-Xyl and Xyl4, and the incorporation of radiolabeled xylose onto Xyl4 was detected by counting the radioactivity (CPM) of the reaction products. (A) XylT activity of representative independent transgenic BY2 cell lines. Note that microsomes from cell lines coexpressing PtrGT43B and PtrGT43C (PtrGT43B/C) exhibited a high level of XylT activity close to that of microsomes from poplar stems, whereas those from lines transfected with the empty vector (control), PtrGT43B alone, or PtrGT43C alone only showed a background level of activity. (B) The XylT activity exhibited by microsomes of the PtrGT43B/C-coexpressing transgenic cell line (PtrGT43B/C-2) is dependent of the Xyl4 acceptor concentration.
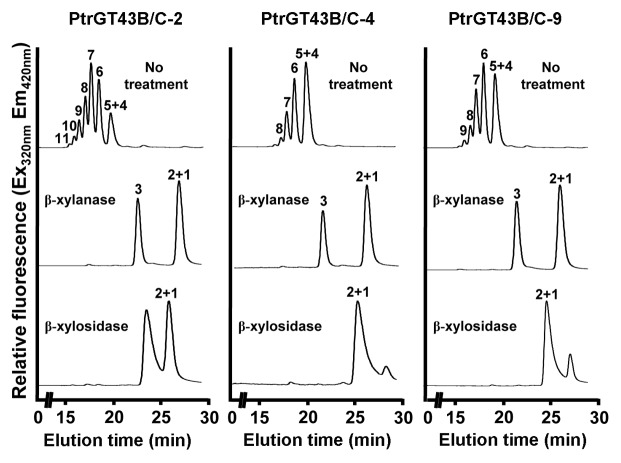
Using fluorescent Xyl4-AA as an acceptor, we revealed that coexpression of PtrGT43B and PtrGT43C led to the successive transfer of multiple xylosyl residues onto Xyl4-AA (Fig. 4). In particular, one of the transgenic lines, PtrGT43B/C-2, was able to incorporate up to 7 xylosyl residues onto Xyl4-AA thus generating xylooligosaccharides with up to 11 xylosyl residues, which is equivalent to the XylT acitivty of poplar stem microsomes. In contrast, only a low-level of incorporation of one to two xylosyl residues was observed in the control. A slight increase in the intensity of xylooligomers with one added xylosyl residue was seen in the transgenic lines expressing PtrGT43B or PtrGT43C alone compared with the control (Fig. 4). The incorporation of multiple xylosyl residues onto Xyl4-AA by PtrGT43B/C was further determined by matrix-assisted laser desorption ionization-time-of-flight-mass spectrometry (MALDI-TOF-MS) showing that in addition to the Xyl4-AA acceptor with a molecular mass of 690, a series of additional masses (m/z 822, 954, 1086, 1218, 1350 and 1482) that differ by an incremental mass of 132 Da were present (Fig. 5). This incremental mass of 132 Da is consistent with the predicted sequential addition of xylosyl residues, each of which has a mass of 132 Da, onto the Xyl4-AA acceptor. Further analysis of the reaction products by treatment with endo-β-(1,4)-xylanase and exo-β-(1,4)-xylosidase demonstrated that the xylooligosaccharides were hydrolyzed by these enzymes yielding di- or tri-xylooligomers (Fig. 6), indicating that the incorporated xylosyl residues in the PtrGT43B/C-catalyzed products are β-(1,4)-linked. Together, these results demonstrate that PtrGT43B and PtrGT43C function cooperatively to transfer multiple β-(1,4)-linked xylosyl residues onto xylooligomer acceptors.
Figure 4.
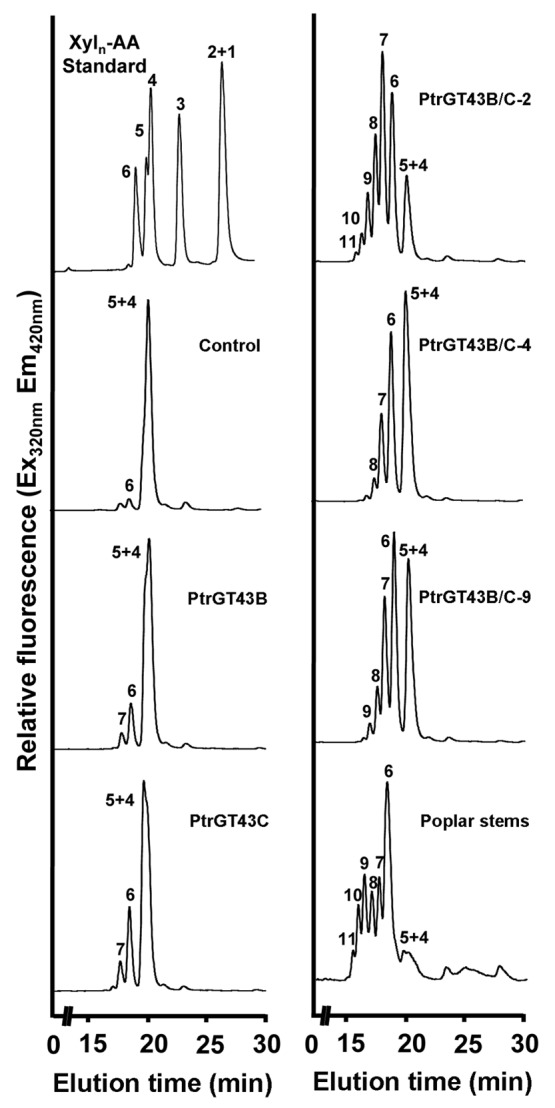
Transfer of multiple xylosyl residues onto the Xyl4-AA acceptor by microsomes with coexpression of PtrGT43B and PtrGT43C. Microsomes from transgenic BY2 cell lines transfected with the empty vector (control), PtrGT43B alone, PtrGT43C alone, or PtrGT43B and PtrGT43C together (PtrGT43B/C-2, -4 and -9) were incubated with UDP-Xyl and the fluorescent Xyl4-AA acceptor, and the reaction products were analyzed by reverse-phase HPLC coupled with fluorescent detection. Note that the XylT activity in transgenic cell lines coexpressing PtrGT43B/C is able to successively elongate Xyl4 to up to Xyl11. The XylT activity of poplar stem microsomes is shown for comparison and a chromatogram of standard Xyl1–6-AA is shown at the top left for reference.
Figure 5.
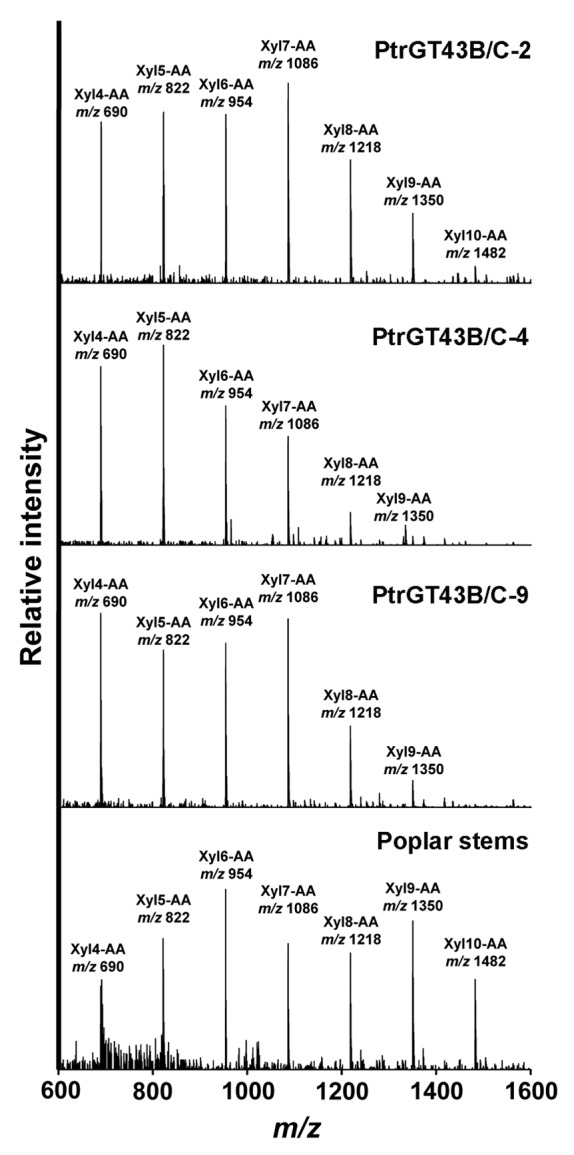
MALDI TOF mass spectra of the PtrGT43B/C-catalyzed reaction products. Microsomes from PtrGT43B/C-coexpressing transgenic cell lines (PtrGT43B/C-2, -4 and -9) and poplar stems were incubated with UDP-Xyl and the Xyl4-AA acceptor, and the reaction products were detected by MALDI TOF MS according to Lee et al.16 The spectra of the reaction products show that in addition to the Xyl4-AA acceptor, a series of ions with a mass increment of 132 D corresponding to one xylosyl residue were present. These ions correspond to AA-labeled oligosaccharides [Xyln-AA + Na]+ composed of 4 to 10 Xyl residues.
Figure 6.
Susceptibility of the PtrGT43B/C-catalyzed reaction products to digestion by endo-β-(1,4)-xylanase and exo-β-(1,4)-xylosidase. Microsomes from PtrGT43B/C-coexpressing transgenic cell lines (PtrGT43B/C-2, -4 and -9) were incubated with UDP-Xyl and the fluorescent Xyl4-AA acceptor, and the reaction products were digested with β-(1,4)-xylanase or β-(1,4)-xylosidase and then analyzed by reverse-phase HPLC according to Lee et al.16 Note that the reaction products were digested into Xyll to Xyl3 by these enzymes, indicating that they have β-(1,4)-linkage.
Our finding that coexpression of PtrGT43B and PtrGT43C is sufficient to achieve the successive transfer of multiple xylosyl residues onto xylooligomer acceptors provides further biochemical evidence supporting the hypothesis that vascular plants evolved to recruit family GT43 members as XylTs catalyzing the elongation of the xylan backbone. Xylan, the second most abundant polysaccharide in dicot wood, represents a significant fraction of biomass sugars that can potentially be harnessed for fermentation into biofuels. On the other hand, its interweaving with cellulose and cross-linking with lignin hinder the accessibility of cell walls by cellulase, thus increasing the recalcitrance of biomass to enzymatic digestions into fermentable sugars.28 Therefore, there is a practical need to identify and functionally characterize xylan biosynthetic genes, which can be used as genetic tools for modification of xylan content and structure tailored for biofuel production. Modification of xylan content and structure in poplar has been shown to lead to a positive impact on the reduction of wood biomass recalcitrance,13,27 which has important implications in genetically engineering of wood biomass better suited for biofuel production. Further genetic and biochemical characterization of other wood-associated glycosyltransferases involved in xylan biosynthesis will promise to provide additional tools for this endeavor.
Acknowledgments
This work was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02-03ER15415.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/19269
References
- 1.Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leplé JC, et al. Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol. 2002;20:607–12. doi: 10.1038/nbt0602-607. [DOI] [PubMed] [Google Scholar]
- 2.Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165–82. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 3.Demura T, Ye Z-H. Regulation of plant biomass production. Curr Opin Plant Biol. 2010;13:299–304. doi: 10.1016/j.pbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Joshi CP, Thammannagowda S, Fujino T, Gou JQ, Avci U, Haigler CH, et al. Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen. Mol Plant. 2011;4:331–45. doi: 10.1093/mp/ssq081. [DOI] [PubMed] [Google Scholar]
- 5.Timell TE. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol. 1967;1:45–70. doi: 10.1007/BF00592255. [DOI] [Google Scholar]
- 6.Suzuki S, Li L, Sun Y-H, Chiang VL. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006;142:1233–45. doi: 10.1104/pp.106.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liepman AH, Nairn CJ, Willats WG, Sørensen I, Roberts AW, Keegstra K. Functional genomic analysis supports conservation of function among cellulose synthase-like a gene family members and suggests diverse roles of mannans in plants. Plant Physiol. 2007;143:1881–93. doi: 10.1104/pp.106.093989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs A, Dahlman O. Characterization of the molar masses of hemicelluloses from wood and pulps employing size exclusion chromatography and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Biomacromolecules. 2001;2:894–905. doi: 10.1021/bm010050b. [DOI] [PubMed] [Google Scholar]
- 9.Shimizu K, Ishihara M, Ishihara T. Hemicellulases of brown rotting fungus, Tyromyces palustris. II. The oligosaccharides from the hydrolysate of a hardwood xylan by the intracellular xylanase. Mokuzai Gakkaishi. 1976;22:618–25. [Google Scholar]
- 10.Johansson MH, Samuelson O. Reducing end groups in birch xylan and their alkaline degradation. Wood Sci Technol. 1977;11:251–63. doi: 10.1007/BF00356924. [DOI] [Google Scholar]
- 11.Andersson S-I, Samuelson O, Ishihara M, Shimizu K. Structure of the reducing end-groups in spruce xylan. Carbohydr Res. 1983;111:283–8. doi: 10.1016/0008-6215(83)88312-8. [DOI] [Google Scholar]
- 12.Peña MJ, Zhong R, Zhou G-K, Richardson EA, O’Neill MA, Darvill AG, et al. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–63. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C, Teng Q, Huang W, Zhong R, Ye Z-H. Down-regulation of PoGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol. 2009;50:1075–89. doi: 10.1093/pcp/pcp060. [DOI] [PubMed] [Google Scholar]
- 14.Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, et al. Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 2005;137:983–97. doi: 10.1104/pp.104.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, et al. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52:1154–68. doi: 10.1111/j.1365-313X.2007.03307.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee C, O’Neill MA, Tsumuraya Y, Darvill AG, Ye Z-H. The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol. 2007;48:1624–34. doi: 10.1093/pcp/pcm135. [DOI] [PubMed] [Google Scholar]
- 17.Lee C, Teng Q, Huang W, Zhong R, Ye Z-H. The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol. 2010;153:526–41. doi: 10.1104/pp.110.155309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu AM, Hörnblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, et al. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 2010;153:542–54. doi: 10.1104/pp.110.154971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou GK, Zhong R, Himmelsbach DS, McPhail BT, Ye ZH, Ye Z-H. Molecular characterization of PoGT8D and PoGT43B, two secondary wall-associated glycosyltransferases in poplar. Plant Cell Physiol. 2007;48:689–99. doi: 10.1093/pcp/pcm037. [DOI] [PubMed] [Google Scholar]
- 20.Lee C, Teng Q, Zhong R, Ye Z-H. Molecular dissection of xylan biosynthesis during wood formation in poplar. Mol Plant. 2011;4:730–47. doi: 10.1093/mp/ssr035. [DOI] [PubMed] [Google Scholar]
- 21.Zhong R, Peña MJ, Zhou G-K, Nairn CJ, Wood-Jones A, Richardson EA, et al. Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell. 2005;17:3390–408. doi: 10.1105/tpc.105.035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C, Zhong R, Richardson EA, Himmelsbach DS, McPhail BT, Ye Z-H. The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 2007;48:1659–72. doi: 10.1093/pcp/pcm155. [DOI] [PubMed] [Google Scholar]
- 23.Zhou GK, Zhong R, Richardson EA, Morrison WH, 3rd, Nairn CJ, Wood-Jones A, et al. The poplar glycosyltransferase GT47C is functionally conserved with Arabidopsis Fragile fiber8. Plant Cell Physiol. 2006;47:1229–40. doi: 10.1093/pcp/pcj093. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, Teng Q, Huang W, Zhong R, Ye Z-H. The poplar GT8E and GT8F glycosyltransferases are functional orthologs of Arabidopsis PARVUS involved in glucuronoxylan biosynthesis. Plant Cell Physiol. 2009;50:1982–7. doi: 10.1093/pcp/pcp131. [DOI] [PubMed] [Google Scholar]
- 25.Richmond TA, Somerville CR. Integrative approaches to determining Csl function. Plant Mol Biol. 2001;47:131–43. doi: 10.1023/A:1010627314782. [DOI] [PubMed] [Google Scholar]
- 26.Sandhu AP, Randhawa GS, Dhugga KS. Plant cell wall matrix polysaccharide biosynthesis. Mol Plant. 2009;2:840–50. doi: 10.1093/mp/ssp056. [DOI] [PubMed] [Google Scholar]
- 27.Lee C, Zhong R, Ye Z-H. Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol. 2012;53:135–43. doi: 10.1093/pcp/pcr158. [DOI] [PubMed] [Google Scholar]
- 28.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–7. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]