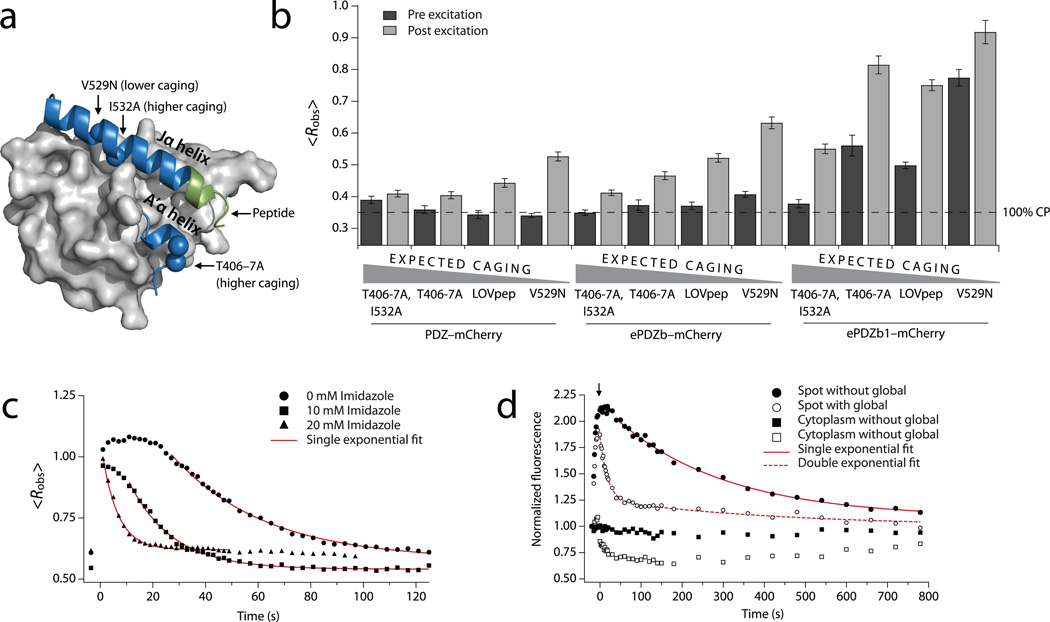
Figure 2.
Mutational and chemical control of binding. (a) AsLOV2 structure (Protein Data Bank: 2v0u) showing the location of the ePDZ epitope (green) and the caging mutations used in this study. (b) Lit- and dark-state <Robs> using LOVpep with caging mutations. Data are the means from a population (n ≥ 34) of cells; error bars show s.e.m. The dashed line represents <Robs> for ~100% cytoplasmic ePDZ–mCherry. (c) Kinetics of global recruitment and dissociation of ePDZb1–mCherry for LOVpep with wild-type dark-recovery kinetics. Imidazole is added to the media in the concentrations indicated. Data are the means from a population (n ≥ 8) of cells. Red lines are exponential fits of the dissociation phase (kobs). (d) Kinetics of spot recruitment and dissociation of ePDZb1–mCherry using slow-cycling (V416I) LOVpep. ePDZb1–mCherry is recruited to a spot as in Fig. 1c. For the filled symbols, the recruited molecules are allowed to recover without further illumination. For the open symbols, the cell is globally photoexcited at the time indicated by the arrow so as to deplete the unbound cytoplasmic pool (open squares) of ePDZb1–mCherry. Data are the means from a population (n ≥ 13) of cells. Red lines are exponential fits of the dissociation phase.