Abstract
Mutant huntingtin (HTT) protein causes Huntington’s Disease (HD), an incurable neurological disorder. Silencing mutant HTT using nucleic acids would eliminate the root cause of HD. Developing nucleic acid drugs is challenging, and an ideal clinical approach to gene silencing would combine the simplicity of single-stranded antisense oligonucleotides with the efficiency of RNAi. Here we describe RNAi by single-stranded silencing RNAs (ss-siRNAs). ss-siRNAs are potent (>100-fold more than unmodified RNA) and allele-selective (>30-fold) inhibitors of mutant HTT expression in cells derived from HD patients. Strategic placement of mismatched bases mimics micro-RNA recognition and optimizes discrimination between mutant and wild-type alleles. ss-siRNAs require argonaute protein and function through the RNAi pathway. Intraventricular infusion of ss-siRNA produced selective silencing of the mutant HTT allele throughout the brain in a mouse HD model. These data demonstrate that chemically modified ss-siRNAs function through the RNAi pathway and provide allele-selective compounds for clinical development.
INTRODUCTION
Huntington’s Disease (HD) is an incurable neurological disorder that afflicts at least 1:100,000 people worldwide (Walker, 2007; Finkbeiner 2011). The disease is characterized by progressive neurodegeneration and symptoms worsen steadily until death. HD is caused by a dominant heterozygous expansion of CAG trinucleotide repeats within the protein-encoding region of the huntingtin (HTT) gene. CAG is the codon for glutamine and the average mutant HTT allele in patients contains approximately 45 consecutive CAG trinucleotides (MacDonald et al, 1993; Duyao et al, 1993; Kremer et al, 1994). While the genetic origin of HD has been known for almost twenty years, curative drugs have not been identified. Effective agents that will benefit HD patients are urgently needed.
HTT is a difficult target for traditional small molecule drugs because it forms interactions with many other proteins and because it is difficult to design small molecules that potently and selectively disrupt protein:protein interactions. Because the genetic origin of HD is localized to just one gene, inhibiting expression of HTT is a promising therapeutic option. Approaches to blocking HTT expression include use of single-stranded antisense oligonucleotides (ASOs) and duplex RNAs (dsRNAs) that target HTT mRNA (Sah and Aronin, 2011; Matsui and Corey, 2012). ASOs and dsRNAs that inhibit expression of HTT have been shown to alleviate symptoms and prolong survival in mouse HD models (Harper et al. 2005; DiFiglia et al, 2007; Boudreau et al., 2009; Drouet et al., 2009), with transient infusion yielding a sustained reversal of phenotype that persists longer than the HTT knockdown (Kordasiewicz et al., 2012). This success suggests that silencing HTT expression can be a productive strategy for developing drugs to treat HD.
HD is dominantly inherited, with patients expressing both mutant and wild-type HTT alleles. Simultaneously inhibiting both alleles may prove to be a successful clinical strategy and studies in a mouse model have shown that reduction of both wild-type and mutant HTT has the same benefit as reduction of mutant HTT alone (Kordasiewicz et al., 2012). Multiple studies, however, suggest that reducing wild-type HTT levels may have deleterious effects (Nasir et al., 1995; Zeitlin et al., 1995; White et al., 1997; Godin et al. 2010; Omi et al, 2005; Huang et al., 2011). Allele-selective inhibitors that maximize reduction of mutant HTT and minimize loss of wild-type HTT would be ideal. One approach to achieving this goal exploits the existence of single nucleotide polymorphisms (SNPs) that allow dsRNAs to distinguish the mutant and wild-type alleles (Miller et al, 2003; Schwarz et al, 2006; van Bilsen et al, 2008; Carroll et al, 2011). The identity of SNPs varies between patients, but some SNPs are common and a few SNPs may be sufficient to cover a majority of HD patients in certain populations (Pfister et al, 2009; Lombardi et al, 2009; Carroll et al, 2011: Warby et al., 2009).
An alternative strategy for allele-selective inhibition exploits a universal difference between the mutant and wild-type alleles – the mutant alleles have more trinucleotide repeats. The longer poly-CAG tract in mutant HTT mRNA offers more binding sites for complementary oligomers. In addition, trinucleotide repeats can form hairpin self-structures (Michlewski and Krzyzosiak, 2004; de Mezer et al, 2011; Krzyzosiak et al, 2011), and the expanded mutant repeats will likely form structures that differ from wild-type. These mutant structures may be more susceptible to recognition and selective binding by oligonucleotides and allow preferential inhibition of the mutant allele. We initially used single stranded ASOs to test the hypothesis that oligomers complementary to CAG repeats could be allele-selective inhibitors (Hu et al., 2009; Gagnon et al., 2010; Hu et al. 2011; Gagnon et al., 2011). We identified several allele-selective ASOs, but did not achieve selectivities of greater than 4–8 fold.
The mechanism of RNAi differs from that of ASOs (Watts and Corey, 2012), and we reasoned that changing our silencing strategy to RNAi might improve selectivities. Our initial tests with fully complementary siRNAs generated potent inhibition but little selectivity (Hu et al., 2009). Fully complementary duplexes function through a siRNA pathway that involves cleavage of target mRNAs while mismatch-containing duplexes can act through a micro RNA (miRNA)-like pathway that suppresses translation (Filipowicz et al., 2008). To test whether duplexes that resemble miRNAs might afford greater selectivity, we altered the mechanism of gene silencing by generating duplexes that mimicked miRNAs by introducing mismatches into the central region of the dsRNA. The mismatches were at positions predicted to disrupt cleavage of the target by argonaute 2 (AGO2) (Wang et al., 2008), an essential protein for substrate recognition and degradation during RNAi (Liu et al., 2004). Using this strategy, we identified mismatch-containing RNAs that were potent and selective inhibitors (Hu et al., 2010). Krzyzosiak and colleagues also reported allele-selectivity using RNA duplexes with mismatches at positions 13 and 16 (Fiszer et al., 2011).
Compounds that combine the favorable biodistribution and simpler synthesis of single-stranded oligonucleotides with the potency of duplex RNAs would offer an ideal strategy for silencing gene expression. Single-stranded RNAs (ssRNAs) has been reported to enter the RNA induced silencing complex (RISC) under specific conditions and inhibit gene expression (Martinez et al., 2002; Schwarz et al. 2002; Holen et al., 2003). Unlike duplex RNA, which is stable in serum, the half-life of ssRNA in serum is measured in seconds to minutes (Braasch et al., 2003). Most ssRNAs would be likely degraded by nucleases before entering cells and inhibiting gene expression. Chemical modifications can stabilize RNA, and one study has reported that chemically modified boranophosphate RNA single strands could be active inside cells (Hall et al., 2006). There was no experimental follow-up and little exploration of mechanism for unmodified or modified single-stranded RNAs, leaving it unclear whether the approach could have practical application. Another report of gene silencing by chemically modified ssRNAs has appeared (Haringsma et al., 2012), but their robustness and mechanism of action remains unclear.
Recently, the action of ssRNAs that function through the RNAi pathway has been revisited (Lima et al., 2012). Systematic chemically modifications and iterative design improvements led to stabilized single-strand silencing RNAs (ss-siRNAs) (Figure 1A) that efficiently enter the RNA pathway and silence gene expression. Here we describe potent and allele-selective inhibition of mutant HTT expression in HD patient-derived cells and HD model mice ss-siRNAs targeting CAG repeats.
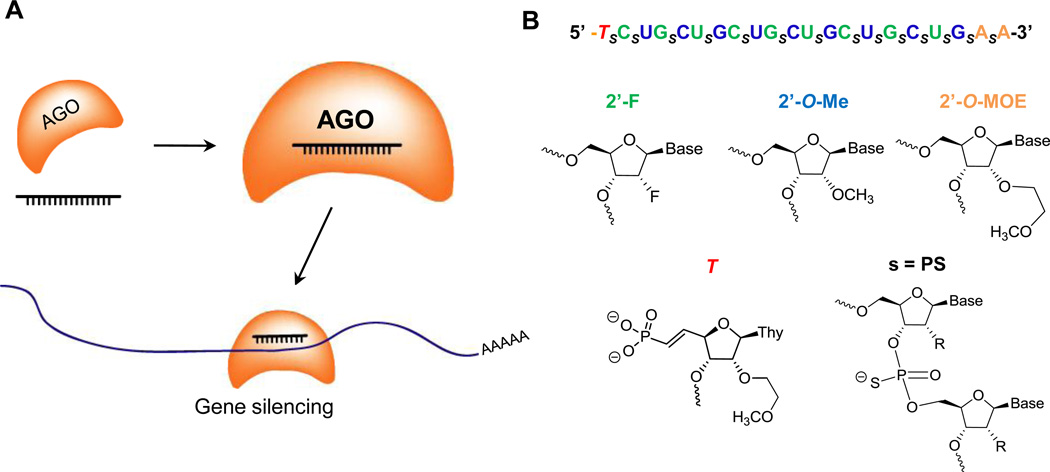
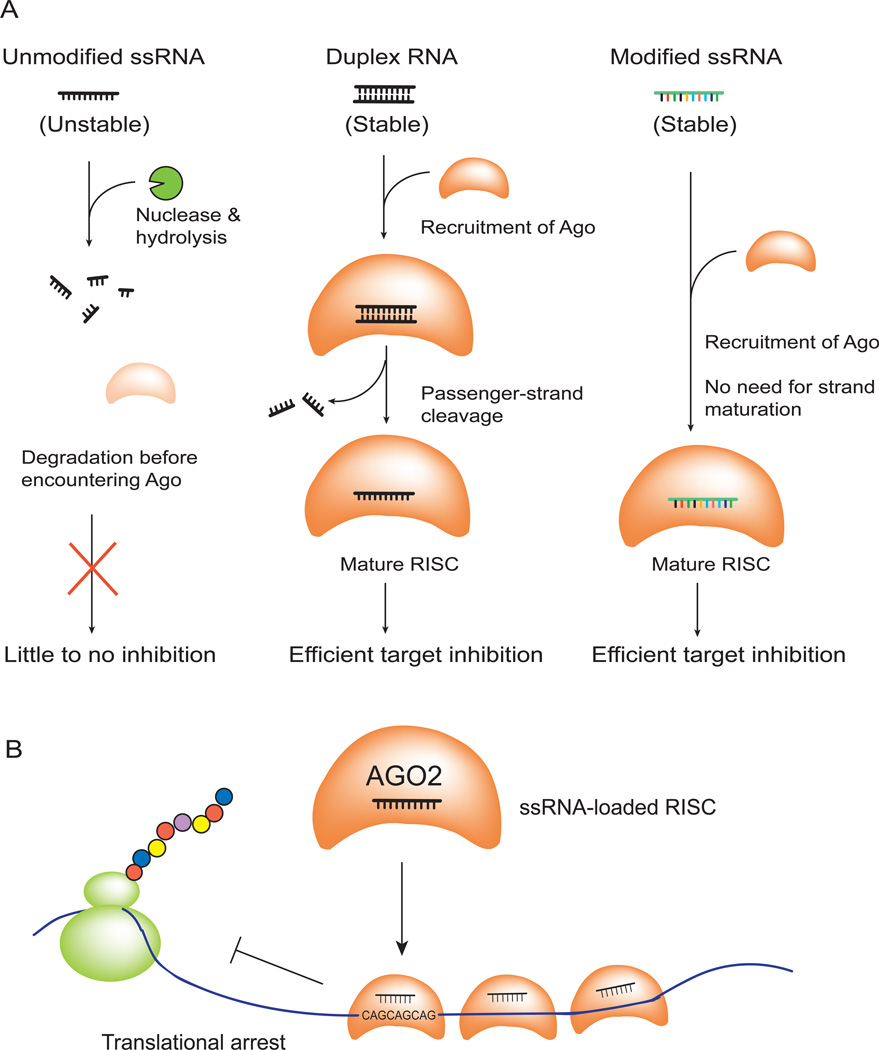
Figure 1. AGO-modulated gene silencing and chemically modified ss-siRNAs.
(A) Recognition of mRNA by ss-siRNA. ss-siRNA loads into AGO protein and the complex recognizes a target sequence within an mRNA to silence gene expression.
(B) Sequence and chemical modifications of a typical ss-siRNA. The 5'-thymidine base is modified with an (E)-vinylphosphonate.
RESULTS
ss-siRNAs are potent and allele-selective inhibitors of mutant HTT expression
The ss-siRNAs used in these studies contain a mixture of 2’-fluoro (2'-F), 2’-O-methyl (2'-OMe), and 2’-methoxyethyl (2'-MOE) ribose modifications (Figure 1B). The ss-siRNAs possess both phosphodiester and phosphorothioate internucleotide linkages. The 5' terminus was capped with either a phosphate or a 5'-(E)-vinylphosphonate. We initially tested ss-siRNAs 537787 (fully complementary to the CAG repeat) and ss-siRNAs 537775 and 537786 (containing mismatch bases at positions 9 (P9) or 10 (P10) respectively).
These ss-siRNAs were introduced into HD patient derived fibroblast cell line GM04281 (69 CAG repeats/mutant allele, 17 CAG repeats/wild-type allele) by standard transfection methods. From our previous studies, oligomers with centrally located mismatches relative to their mRNA targets are predicted to inhibit expression of HTT protein but have little effect on HTT mRNA (Hu et al., 2010), leading us to focus on measuring protein levels. HTT is a large protein, 347 kDa in molecular weight and the expanded repeat leads to only a few kDa increase. The small molecular weight difference makes discrimination between alleles challenging, but mutant and wild-type alleles can be efficiently resolved using temperaturecontrolled SDS-PAGE (Hu et al., 2009).
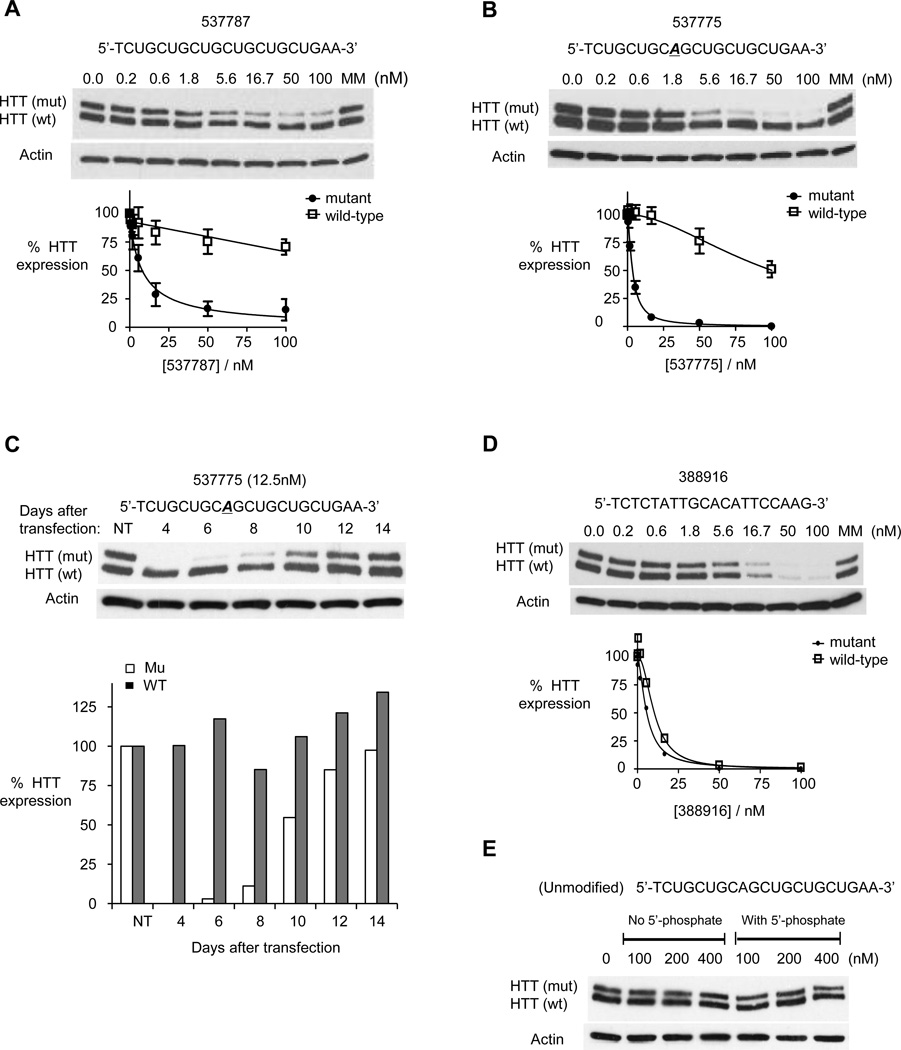
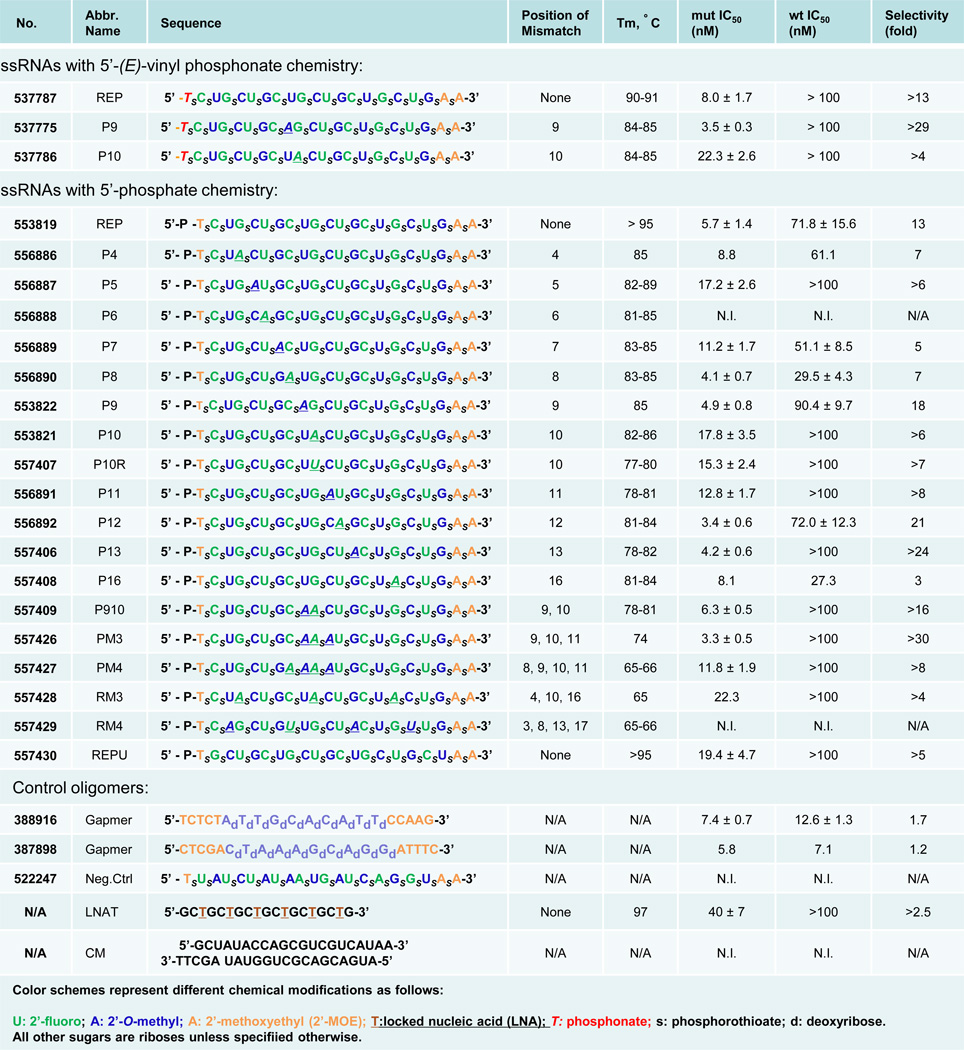
ss-siRNAs 537787, 537775, and 537786 inhibited HTT expression with varying potencies and selectivities (Figure 2 and Figure 3). Fully complementary ss-siRNA 537787 possessed an IC50 value of 8 nM and a selectivity of >13 fold (Figure 2A). This selectivity is better than the selectivity of the analogous unmodified duplex RNA (2-fold) (Hu et al. 2009). ss-siRNA 537775 (single mismatch at position 9) possessed an IC50 value of 3.5 nM and a selectivity of >29 fold (Figure 2B). ss-siRNA 537775 was the best inhibitor with potency and selectivity values similar to the most selective dsRNAs identified (Hu et al., 2010). ss-siRNA 537786 (single mismatch to P10, one base shift relative to 537775) possessed an IC50 value of 22.3 nM and a selectivity of >4 fold (Figure 3), making it less effective than the analogous mismatch-containing dsRNA (Hu et al., 2010).
Figure 2. ss-siRNAs inhibit HTT expression.
Western analysis of inhibition of HTT expression by:
(A) ss-siRNA 537787 (no mismatches);
(B) ss-siRNA 537775 (one mismatch at P9).
(C) ss-siRNA 537775 over fourteen days with quantitation.
(D) a methoxyethyl antisense oligonucleotide that targets a non-repeat region of HTT mRNA. The graphs in parts A,B, and D show amounts of both wild-type and mutant HTT protein.
(E) a fully complementary single-stranded RNA lacking any chemical modifications with and without a 5’ terminal phosphate.
MM: an RNA duplex containing multiple mismatches. Western analysis is representative data of at least duplicate experiments. Error bars are standard error of the mean (SEM) for dose response studies from three or more independent experiments.
Figure 3. ss-siRNAs and other oligonucleotides used in these studies.
N.I.: no inhibition; N/A: Not available. Except for CM (a dsRNA species), all Tm's are measured using ss-siRNAs duplexed with equimolar amounts of unmodified ssRNA 5'- CAGCAGCAGCAGCAGCAGCAGC-3'. Gapmer oligonucleotides have backbones containing only phosphorothioate linkages. All data were obtained using HD patient-derived fibroblast cell line GM04281. Selectivity is calculated by dividing the IC50 of for inhibition of wild-type HTT expression by that for mutant HTT. Error ranges represent standard error of the mean (SEM) of IC50 values from biological replicates.
These results suggest that chemical modifications and the precise positioning of mismatched bases affect allele-selective recognition and silencing. In one case selectivity was better relative to the analogous dsRNA, in another selectivity was similar, and in the third example selectivity and potency were worse. While ss-siRNAs appear to function similarly to dsRNAs, the exact outcome of recognition depends on sequence.
We examined inhibition of HTT by ss-siRNA 537775 over time. ss-siRNA was added only once, at the beginning of the experiment. During this period, cells doubled 3–4 times, diluting out the ss-siRNA. We observed >80% inhibition of mutant HTT expression for up to 8 days, with expression gradually returning to the original levels after two weeks (Figure 2C).
To provide a comparison with a silencing strategy that is non-allele selective and does not involve RNAi, we tested a gapmer ASO complementary to a region outside the CAG tract. The “gapmer” ASO contains a central DNA “gap” to recruit RNase H that is flanked by chemically modified bases to improve binding (Watts et al., 2012). The gapmer inhibited HTT expression with an IC50 value of 7.4 nM and a selectivity of 1.7-fold (Figure 2D and Figure 3). These data suggest that ss-siRNAs targeted to the CAG repeat can achieve potencies that are similar to those achieved using ASOs, a gene silencing approach broadly used in clinical testing (Watts and Corey, 2012) while having the added benefit of being allele-selective.
As a further comparison, we examined silencing by anti-CAG ssRNAs that lacked chemical modifications. These unmodified ssRNAs were not active when tested at concentrations of up to 400 nM (Figure 2E). These results contrast with reports that unmodified ssRNAs possessed silencing activity in mammalian cells (Martinez et al., 2002; Schwarz et al, 2002) possibly due to differences in the cell lines, transfection techniques, the mRNA target, or the sequence of the silencing RNA.
Optimizing ss-siRNA design
ss-siRNAs 537787, 537775, and 537786 contain 5’-terminal vinylphosphonate moieties designed to improve stability and potency in vivo. This modification is not needed for testing in cell culture (Lima et al., 2012) and substitution with a phosphate moiety facilitates the synthesis of the large number of compounds needed to identify improved inhibitors and investigate mechanism. To determine whether potent and allele selective inhibition could be achieved with 5’-phosphate ss-siRNAs, we tested compounds 553819, 553822, and 553821. These ss-siRNAs possessed potencies and selectivities similar to their phosphonate analogues (Figure 3 and Figure 4A, Figure S1).
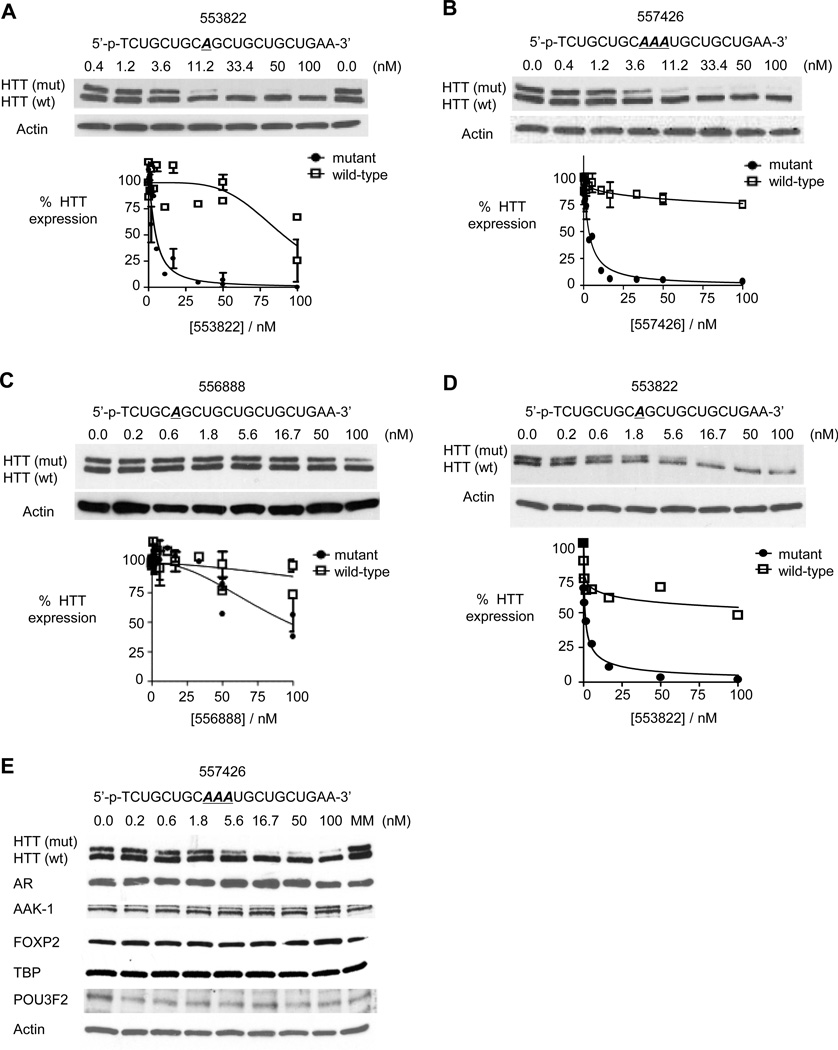
Figure 4. Characterization of inhibition by modified ss-siRNAs.
Western analysis of inhibition of HTT expression by:
(A) ss-siRNA 553822 (mismatched base at P9) containing a 5'-phosphate;
(B) ss-siRNA 557426 containing three central mismatches;
(C) ss-siRNA 556888 containing a mismatch at P6; and
(D) ss-siRNA 553822 (mismatched base at P9) in 44-CAG-repeat GM04719 cells.
(E) Effect of ss-siRNA 537775 on other genes containing trinucleotide repeats.
MM: an RNA containing multiple mismatches. Western analysis for parts A–C is representative data from three or more experiments and error bars are standard error of the mean (SEM).
We chose the phosphate design for large-scale tests and synthesized compounds that varied in the number and placement of mismatched bases (Figure 3). Several compounds possessed good potencies and selectivities, with the best combination of potency and selectivity achieved by ss-siRNA 557426. ss-siRNA 557426 contained three centrally located mismatched bases and combined an IC50 value of 3.3 nM with >30 fold selectivity (Figure 3 and Figure 4B).
We had previously observed that a mismatched base at position P6 within a dsRNA abolished RNA-mediated inhibition of HTT. P6 is within the seed sequence (bases 2–8), a region critical for efficient RNAi (Lim et al., 2005). ss-siRNA 556888, an ss-siRNA that contained a mismatched base at P6, was the only single-mismatch compound to not inhibit HTT (Figure 3 and Figure 4C). This result suggests that ss-siRNAs and duplex RNAs share critical recognition elements and supports the hypothesis that ss-siRNAs act through the RNAi pathway.
Most HD patients have mutant HTT alleles containing 40–50 repeats (Duyao et al., 1993; MacDonald et al. 1993). To determine whether ss-siRNAs might also be an effective strategy for allele-selective inhibition in this patient cohort, we tested inhibition in GM04719 patient derived fibroblast cells (44 mutant repeats/15 wild-type repeats) (Figure 4D). We found that phosphate ss-siRNA 553822 inhibited expression of mutant HTT with an IC50 value of 0.9 nM and an allele-selectivity >100-fold, demonstrating the potential to achieve allele-selectivity in cell lines within the median range of CAG repeat copy number. We note that inhibition of wild-type HTT levels off at approximately 45%. This suggests a population of wildtype mRNA that contains some species that are refractory to silencing by anti-CAG ss-siRNAs.
Another challenge for agents that target trinucleotide repeats is the existence of other genes that contain repetitive regions (Kozlowski et al., 2010). We observed no inhibition of TATA-box binding protein (19 CAG repeats), androgen receptor (~20 CAG repeats), AAK-1 (6 CAG repeats), POU3F2 (6 CAG repeats), or FOXP2 (40 glutamines encoded by a mix of CAG and CAA trinucleotides) (Figure 4E) at concentrations well above those needed to achieve selective inhibition of mutant HTT.
Involvement of AGO2 protein
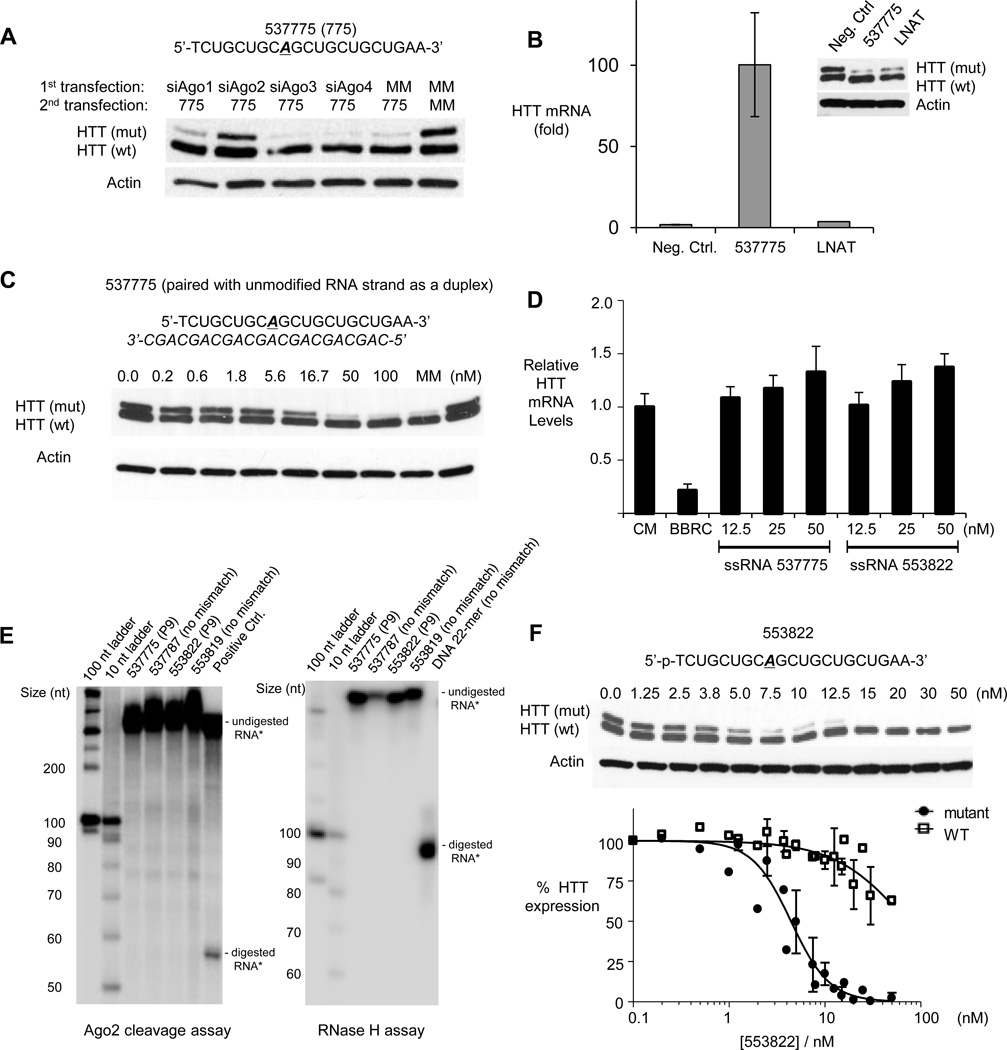
AGO2 is a key protein involved in RNAi (Liu et al., 2004; Meister et al., 2004). There are four AGO genes in human cells. AGO2 is the best characterized and the only variant with endonucleolytic activity. AGO1, AGO3, and AGO4 are also expressed, but their functions are less well defined. To determine which AGO variant is involved in gene silencing by ss-siRNAs, we used siRNAs targeting mRNAs encoding AGO1-4 to reduce expression (Figure S2). We observed that reducing AGO2 levels reversed silencing by 537775, consistent with involvement of AGO2 (Figure 5A). By contrast, silencing AGO1, AGO3, or AGO4 had little effect on allele-selective inhibition of mutant HTT.
Figure 5. Mechanism of allele-selective inhibition of HTT by ss-siRNA.
(A) Western analysis of the effect siRNA-mediated reduction of AGO1-4 expression on allele-selective inhibition by ss-siRNA 537775.
(B) RNA immunoprecipitation (RIP) using anti-AGO2 antibody after transfection of ss-siRNA 537775, a control ss-siRNA 522247 not targeting HTT, or an allele-selective single-stranded ASO (LNAT) (Hu et al., 2009) at 25nM. Y-axis measures fold-enrichment of HTT mRNA of anti-AGO2 vs. IgG pulldown.
(C) Western analysis of inhibition of HTT expression by ss-siRNA 537775 in complex with a complementary unmodified RNA.
(D) Effect of ss-siRNAs 537775 or 553822 on levels of HTT mRNA evaluated by QPCR.
(E) In vitro assays using recombinant RNase H and Ago2 proteins do not show efficient substrate cleavage by ss-siRNA.
(F) Primary data and Hill plot used for determining cooperativity of HTT inhibition by ss-siRNA 553822. X-axis shows ss-siRNA concentration in logarithmic scale. Hill’s coefficient (nh) is 2.2 ± 0.3 for mutant HTT and 1.2 ± 0.2 for wild-type HTT.
Error bars from western quantitation and RIP are standard error of the mean (SEM) from three or more independent experiments, and error bars on HTT mRNA levels are standard deviations (SD) from replicate data.
See also Figure S2.
To further investigate involvement of AGO, we used RNA immunoprecipitation (RIP) to examine the ability of ss-siRNA 537775 (P9 mismatch, phosphonate 5’ terminus) to promote association of AGO2 with HTT mRNA. We transfected ss-siRNA 537775 into cells, harvested extracts, immunoprecipitated AGO-bound material using an anti-AGO2 antibody, and assayed the abundance of HTT mRNA.
We observed that HTT mRNA could be recovered upon transfection of ss-siRNA 537775 and RIP with anti-AGO2 antibody, but not when we used a noncomplementary ss-siRNA (Figure 5B). A locked nucleic acid (LNA) ASO that targets the CAG repeat and inhibits mutant HTT expression with an allele-selectivity >6 fold (Hu et al., 2009; Gagnon et al, 2010) did not recruit AGO2 to HTT mRNA. The difference between the ss-siRNA and the LNA ASO underlines a fundamental difference in the mechanisms of action – ss-siRNAs rely on AGO2 while ASOs do not. Taken together, results from gene silencing and RNA immunoprecipitation support the conclusion that AGO2 is required for the action of ss-siRNA and that silencing proceeds through the endogenous RNAi pathway.
AGO is typically thought to mediate recognition of mRNA inside cells by dsRNA consisting of a guide strand hybridized to a passenger strand. To determine the functional necessity of passenger strand, we created a duplex by annealing ss-siRNA 537775 to an unmodified RNA passenger strand. This heteroduplex inhibited HTT expression with an IC50 of 5.4 nM and a selectivity of >15-fold (Figure 5C), similar to the ss-siRNA alone (IC50: 3.7 nM; selectivity: >29-fold). This result demonstrates that introduction of chemically modified bases into the guide strand does not interfere with strand loading and that ss-siRNA can function through RNAi pathways that in the past were thought to require a passenger strand. This finding is significant because it shows that ss-siRNA is the only active species during gene silencing and the passenger strand is not necessary. During standard dsRNA-mediated RNAi using unmodified RNA, the main role of the passenger strand is likely to protect the guide strand from digestion by nucleases, ensuring that it survives long enough to reach its target mRNA.
Inhibitory ss-siRNAs do not reduce HTT mRNA levels
siRNAs that are fully complementary to their target mRNAs are usually thought to cause AGO2-mediated mRNA cleavage and reduction of mRNA levels. The introduction of centrally located mismatches is predicted to interfere with strand cleavage without affecting binding (Wang et al., 2008). To test this hypothesis, we measured RNA levels by quantitative PCR (QPCR) (Figure 5D). A duplex siRNA that targets a sequence outside of the CAG repeat reduces HTT mRNA levels by >80%. By contrast, phosphonate ss-siRNA 537775 and phosphate ss-siRNA 553822 that contain mismatches at position P9 do not reduce RNA levels. This result is consistent with a mechanism that involves blocking protein translation rather than degradation of mRNA.
We also examined the potential for cleavage using an in vitro assay combining purified AGO2 or RNase H, different ss-siRNAs, and an in vitro-transcribed HTT RNA transcript containing seventeen CAG repeats. ss-siRNAs 537775, 556887, 553822, and 553819, all potent and selective inhibitors inside cells, did not lead to transcript cleavage (Figure 5E). By contrast, a control duplex RNA targeting a non-CAG sequence yielded cleavage products of the expected size. A control DNA oligonucleotide yielded cleavage upon addition of RNase H. These data demonstrate that ss-siRNAs targeting the CAG repeat do not cause RNA cleavage through the RNAi or RNase H pathways.
Inhibition by ss-siRNAs is cooperative
The repetitive region within a mutant HTT mRNA with 69 repeats can bind up to 9–10 twenty-base-long oligomers. A wild-type mRNA with 17 repeats, by contrast, can bind no more than two. It is possible that binding of multiple oligomers at adjacent sites can lead to cooperative inhibition and contribute to allele-selective recognition of expanded mutant repeat regions. To determine whether inhibition is cooperative, we examined inhibition of mutant HTT by ss-siRNA 553822 over a wide range of concentrations (Figure 5F). After fitting the data to the Hill equation we calculated Hill coefficients, nh, of 2.2 and 1.2 for inhibition of mutant and wild-type HTT expression respectively. These data are consistent with cooperative inhibition and suggest that association of the ss-siRNA with the expanded mutant repeat is likely to involve multiple binding events.
An ss-siRNA is an allele-selective inhibitor in HD model mice
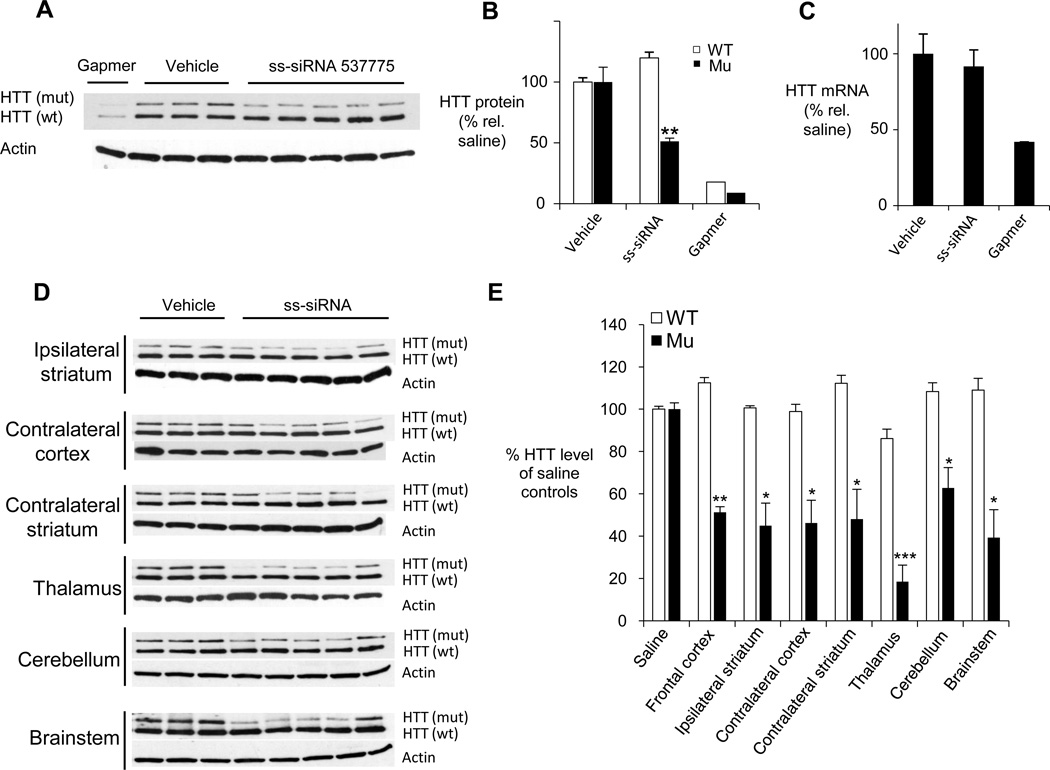
To test ss-siRNAs in animals, we used HdhQ150 heterozygous knock in HD-model mice (Lin et al., 2001). The HdhQ150 heterozygous mice carry one mouse huntingtin allele with 150 CAG repeats knocked into Exon 1 (Q150) and a second allele with a wild-type mouse huntingtin gene (Q7). The two HTT alleles in the HdhQ150/Q7 animals differ only in the length of the CAG repeat, making them ideal for determining whether an ss-siRNA can discriminate between the expanded and unexpanded huntingtin transcripts in vivo.
To best mimic the human treatment paradigm, vinylphosphonate ss-siRNA 537775 was introduced into the cerebral spinal fluid of the right lateral ventricle to achieve distribution throughout the CNS, including brain regions implicated in HD pathology. ss-siRNA 537775 was continuously infused into the right lateral ventricle for twenty-eight days (300 µg/day). Due to the long in vivo half-life of the huntingtin protein and the need to monitor protein levels rather than RNA, animals were treated for four weeks to ensure that reduced huntingtin synthesis could be detected.
We analyzed brain tissue for HTT expression by western analysis and Q-PCR. As a positive control we used a nonallele-selective gapmer ASO complementary to a region outside the CAG repeat (administered at 75 µg/day for fourteen days). We observed allele-selective inhibition of HTT protein expression in the frontal cortex of all five mice in the experimental cohort relative to animals treated with saline (Figure 6AB). Q-PCR showed no decrease in HTT mRNA levels in animals treated with ss-siRNA (Figure 6C), consistent with results in cultured cells showing inhibition does not result from cleavage of mRNA.
Figure 6. Allele-selective inhibition of HTT by ssiRNA in Q150 HD mouse model.
(A) Western analysis of HTT expression on allele-selective inhibition by ss-siRNA 537775 (n=5) in Q150/Q7 mouse frontal cortex.
(B) Quantitation of wild-type and mutant HTT protein levels shown in (A).
(C) Q-PCR analysis of HTT mRNA levels in mouse frontal cortex after treatment with ss-siRNA, vehicle, or control gapmer ASO.
(D) Western analysis HTT expression after allele-selective inhibition by ss-siRNA 537773 (n=5) in different brain regions.
(E) Quantitation of western analysis from (D). Results from each treatment group/brain section were averaged.
Error bars represent standard error of the mean (SEM) after averaging quantitation results from multiple gel images. *p < 0.05; **p < 0.01; ***p < 0.001.
See also Figure S3.
We then assayed inhibition in other tissues, including contralateral cortex, thalamus, ipsilateral striatum, contralateral striatum, cerebellum, and brainstem, all of which displayed a reduction in levels of the mutant HTT protein when treated with ss-siRNA 537775 (Figure 6DE). Consistent with our results in cultured cells, injection of ss-siRNA 537775 did not reduce expression of other proteins containing trinucleotide repeats (Figure S3). These experiments demonstrate that ss-siRNAs can distribute broadly throughout the central nervous system and inhibit mutant HTT expression.
DISCUSSION
Therapeutics that slow or reverse progression of HD are a major unmet medical need. Trinucleotide expansions cause numerous other hereditary diseases (Orr and Zoghbi, 2007), and anti-CAG agents that treat HD might also advance treatments for these conditions. sssiRNAs combine strengths of dsRNAs and ASOs, and our objective for this study was to determine whether they would provide a alternate starting point for HD drug development. Substantial challenges confront the application of gene silencing strategies to neurological disorders (Sah and Aronin, 2011; Davidson and McCray, 2011) and optimizing the chemical properties of inhibitory molecules for maximal biological effect is a central goal. Our results demonstrate that ss-siRNAs can mimic miRNAs to allele-selectively suppress translation and inhibit mutant HTT expression with potencies and allele-selectivities that are at least equal to those possessed by duplex RNAs and ASOs.
More broadly, gene silencing strategies that use synthetic nucleic acids have the potential to provide a new class of clinical agents for treating diseases that are currently incurable or where current therapies are inadequate (Watts and Corey, 2012). ss-siRNAs provide a starting point for drug development and an additional option for overcoming roadblocks to successful clinical application. For basic research, ss-siRNAs provide a fresh perspective on the mechanism of RNAi.
ss-siRNAs function through RNAi
miRNAs function through the RNAi pathway and almost all miRNAs contain mismatched bases relative to their targets. We provide several lines of evidence that mismatch-containing anti- CAG ss-siRNAs also act through RNAi: i) the maximum selectivity of inhibition by ss-siRNAs (> 30 fold) (Figure 3) is much closer to that produced by duplex RNA (> 30 fold) (Hu et al, 2010) than that yielded by the non-RNAi-mediated ASOs (> 4.8-fold) (Hu et al., 2009; Gagnon et al., 2010); ii) reduction of AGO2, a key RNAi factor, leads to less efficient silencing of mutant HTT (Figure 5A); iii) addition of ss-siRNA, but not an allele-selective ASO, leads to robust recruitment of AGO2 to HTT mRNA (Figure 5B); iv) adding an unmodified RNA guide strand to the ss-siRNA does not affect its activity (Figure 5C); and v) as observed for dsRNA (Hu et al., 2010), introduction of a mismatch at position 6 within the putative seed sequence for recognition by ss-siRNA largely abolishes inhibition of HTT (Figure 4C).
Action through the RNAi pathway is accompanied by potent inhibition of HTT expression. Several compounds possess IC50 values less than 10 nM and the best ss-siRNAs have potencies that are almost identical to those observed for duplex RNAs (Hu et al., 2010). Easy identification of multiple potent and selective compounds that function through RNAi also has implications for therapeutic development. It is likely that many other compounds, with different mismatch positions or patterns of chemical modifications, will also be active. This large design-space is compatible with allele-selective inhibition and provides many options for optimizating drug-like characteristics and the subsequent development of therapeutics. If one compound has a toxic effect related to its sequence or chemical composition, numerous other compounds can be developed instead.
While all data indicate that ss-siRNAs function through the RNAi pathway, knowing how a dsRNA functions will not always fully predict the properties of an analogous ss-siRNA. For example, fully complementary ss-siRNAs were allele-selective inhibitors of HTT expression (Figure 2A) while the analogous fully complementary dsRNA was not selective (Hu et al., 2009). In another example, we had previously observed that duplex RNAs with mutations at positions 9 or 10 are equally potent and selective inhibitors (Hu et al., 2010). ss-siRNAs with mutations at positions 9 or 10, by contrast, are quite different in potency (Figure 3).
The origin of functional differences between dsRNAs and ss-siRNAs lies in the chemical differences between modified and unmodified RNA. ss-siRNAs have 2’-F and 2’-O-sugar modifications, as well as phosphorothioate internucleotide linkages. The 2’ modifications tend to increase affinity, while phosphorothioate linkages tend to decrease affinity. The countervailing and sometimes unpredictable effects of these modifications are apparent from our data. Compared to analogous unmodified RNAs, some RNAs have lower melting temperature (Tm) values for association with complementary sequences, while others have higher values. In addition, the extensive chemical modification may affect RNA recognition, AGO loading, and subsequent inhibition of gene expression. Understanding the intrinsic properties and potential of ss-siRNA will be an important goal for future research. Many chemically modified bases exist that can be substituted within ss-siRNAs, and it is likely that chemical optimization of recognition, potency, and selectivity will prove a productive area of investigation.
Mechanism of allele-selective inhibition
We have shown that our anti-CAG ss-siRNAs bind AGO2, suggesting that the first steps of allele-selective inhibition by ss-siRNAs involve recognition of AGO2 and subsequent association with HTT mRNA. Mutant and wild-type HTT mRNAs both contain CAG sequences but the lengths of the repeat regions differ. In our most extreme case, selectivity is achieved even though the difference in the number of repeats in GM04719 cells (44 mutant repeats versus 15 wild type repeats) is only 29. One explanation is that the mutant allele provides more binding sites for the ss-siRNA. For example, a wild-type allele with twenty repeats would have space for no more than three ss-siRNAs, while a mutant allele with 40 repeats would have space for as many six ss-siRNAs. Our observation of cooperative effects on inhibition of HTT expression support the conclusion that multiple ss-siRNAs bind to the expanded repeat target and lead to a preference for the mutant over the wild-type repeat. Indeed, the potential for multiple binding of small RNAs to adjacent sites to lead to cooperative gene silencing has been noted previously using expression constructs containing 3’-untranslated regions with varying numbers of target sequences (Broderick et al., 2011).
Once bound, inhibition leads to potent reduction of mutant HTT protein expression but no change in mRNA levels. Inhibition by ss-siRNAs is much more efficient than by analogous unmodified ssRNAs (Figure 2E) and is as efficient as analogous duplex RNAs (Hu et al., 2010). The guide strand, therefore, appears to be the only strand necessary for efficient RNAi (Figure 7A). For experiments with conventional duplex RNAs, the passenger strand serves as a delivery agent protecting the critical guide strand.
Figure 7. Action of chemically modified ss-siRNAs and allele-selective inhibition of HTT.
(A) Chemical modifications allow ss-siRNA to be stable and function through RNAi pathway inside cells.
(B) Binding of multiple anti-CAG ss-siRNA:AGO2 complexes to expanded trinucleotide repeat contributes to allele-selective inhibition.
For mismatch-containing ss-siRNAs, the observed reduction in protein but not mRNA is consistent with our initial design assumptions that mismatches would disrupt AGO-mediated cleavage of mRNA. After multiple ss-siRNAs bind in complex with AGO to the repeat, they likely act as a roadblock to ribosome progress and prevent protein translation (Figure 7B). This “steric blocking mechanism” is similar to that used by ASOs that lack the ability to recruit RNase H and cannot cause cleavage of mRNA, except that in this case the ss-siRNA is delivered by the endogenous RNAi machinery that has greater potential to facilitate efficient gene silencing. Multiple binding sites within expanded repeats permit cooperative binding and discrimination relative to shorter wild-type repeats.
miRNA verus siRNA mechanisms
Crooke and co-workers also have reported inhibition of gene expression by ss-siRNAs in cell culture and animals (Lima et al., 2012). Both manuscripts demonstrate that the passenger strand is not necessary for gene silencing and dispensing with it has the potential to improve the in vivo pharmacology of compounds that function through the RNAi pathway. However, the mechanisms of action differ. Crooke and colleagues use ss-siRNAs that are fully complementary to their target sequences and would be predicted to function through a siRNA-like pathway. Consistent with this expectation, they observe cleavage of target sequence in vitro, 5'-RACE products consistent with AGO2-mediated cleavage of the predicted target site, and reduction of mRNA in cell culture and in vivo.
By contrast, we use mismatch-containing RNAs that resemble miRNAs and have the potential to act through a miRNA-like pathway. Our target is the expanded CAG repeat within mutant HTT mRNA, which offers multiple sites for binding. Consistent with action through a mechanism that resembles that used by miRNAs, we do not observe cleavage of substrate in vitro nor do we observe reduced mRNA levels in cell culture or animals. The observation of ss-siRNA silencing through both the siRNA and miRNA pathways suggests a broad compatibility with RNAi machinery and cellular RNA targets.
Implications for therapeutic gene silencing
Advances in nucleic acid chemistry, a better understanding of nucleic acid pharmacology, and a more mature appreciation of the basic science underlying disease have led to substantial recent clinical progress for nucleic acid therapeutics (Watts and Corey, 2012). It is now possible to cite several examples of nucleic acid drugs that have potent effects on target gene expression in humans. For example, Mipomersen, a drug designed to treat familial hypocholesterolemia, has been shown to benefit patients in multiple Phase III trials and is now awaiting FDA review. Even brain disorders are becoming more amenable to nucleic acid silencing. ASOs have been shown to inhibit superoxidase dismutase in the spinal cord of primates and a Phase I trial designed to test treatment of patients with familial ALS is ongoing.
Our results introduce ss-siRNAs as an alternate strategy for treating neurodegenerative disease that provides an alternative to ASOs or dsRNAs. Chemically, ss-siRNAs are similar to ASOs because they both possess a single chemically modified antisense strand. Mechanistically, they resemble duplex RNAs that function through RNAi. ss-siRNAs combine strengths of the two existing approaches, possess unique advantages, and provide a distinctive new strategy for silencing gene expression.
Here we demonstrate that the first generations of anti-CAG ss-siRNAs achieve potencies and selectivities for inhibiting HTT that are similar to those achieved by well-established gene silencing technologies. By further optimizing the type of chemical modification, placement of mismatched bases, or other design features it is likely that subsequent generations of inhibitory ss-siRNAs will possess even more favorable properties. The availability of a gene silencing strategy that combines the strengths of siRNAs and ASOs will provide an important option for transforming the potential benefits from nucleic acid silencing into practical benefits for patients.
EXPERIMENTAL PROCEDURES
Cell culture
ss-siRNAs were synthesized by Isis Pharmaceuticals Inc. (Carlsbad, CA, USA) and reconstituted in nuclease-free water. Patient-derived fibroblast cell lines GM04719 (44 CAG repeat) and GM04281 (69 CAG repeat) were obtained from the Coriell Institute (Camden, NJ, USA) and transfected as described (Hu et al., 2010). Cells were plated in 6-well plates at 60,000 cells/well in supplemented MEM media 2 days prior to transfection. 6-well plates were used to provide the number of cells necessary for western analysis. Cells were transfected using lipid RNAiMAX (Invitrogen) per manufacturer instructions. Cells were harvested 4 days after transfection for protein analysis and 3 days after transfection for RNA analysis.
Analysis of HTT protein expression
SDS-PAGE (separating gel: 5% acrylamide-bisacrylamide [50:1], 450mM Tris-acetate pH 8.8; stacking gel 4% acrylamide-bisacrylamide [50:1], 150mM Tris-acetate pH 6.8) was used to separate wild-type and mutant HTT proteins as described (Hu et al. 2010).
Analysis of HTT mRNA expression
Quantitative PCR was performed as described. Experiments were performed in biological triplicate and error reported as standard deviation. The Q-PCR cycles are as follows: 50°C for 2 min; 95°C for 5min; (95°C for 15s; 60°C for 1min) × 40 cycles.
Analysis of AGO2 binding by RNA immunoprecipitation
HD-patient derived GM04281 (69 CAG repeat) fibroblasts were grown in 150 cm2 dishes and transfected with chemically modified ss-siRNA. Cells (~4 × 107) were harvested by trypsin 24 h after transfection in growth media. A small quantity of cells are saved and harvested for protein to check knockdown efficiency by western blot. RNA immunoprecipitation was performed as described using 4 µg anti-AGO2 (4G8, 011-22033, Wako) antibody, or 4 µg normal mouse IgG (12–371, Millipore, for IP) antibody in 0.75mL of IP buffer at 4°C on rotator for 3–4 h. Results were normalized by the two following parameters: (i) ratios of HTT mRNA to GAPDH mRNA (a housekeeping control) to eliminate small variations of total RNA across all samples; (ii) binding of HTT mRNA to anti-Ago2 antibodies over that of IgG to measure fold-enrichment of HTT mRNA in Ago2 IP relative to the non-specific IgG background binding.
IC50, selectivity, and cooperativity calculations
Protein bands were quantified from autoradiographs using ImageJ software. The percentage of inhibition was calculated as a relative value to a no-treatment control sample. The program GraphPad Prism 4 was used to draw the fitting curves for dose response experiments. The Hill Equation was used for fitting in the following form: Y=100[(1−Xn/(Kn+Xn)], where Y is percentage of inhibition, X is the ss-siRNA concentration, K is the IC50 value, and n is the Hill coefficient. At least three experiment data sets were used for curve fitting. The error is standard deviation, calculated from each individual dose curve. Selectivity was calculated by taking the ratio of the IC50 for inhibition of the mutant HTT protein over that of the wild-type protein. Cooperativity was measured by obtaining the Hill’s coefficient that best fits the plotted curve, which corresponds to the value n in the equation.
In vitro Ago2 activity assay
17-CAG RNA was then dephosphorylated in a 40 µL reaction containing 10 U of by calf intestinal phosphatase (CIP) (New England Biolabs), 1× CIP buffer and 20 U of SUPERase-In RNase Inhibitor (Ambion) for 15 min at 37°C then 15 min at 55°C. RNA was extracted with pH 4.3 phenol and 24:1 chloroform:isoamyl alcohol then precipitated with 0.3 M sodium acetate and 70 % ethanol.
Dosing and surgical procedure
HdhQ150 (CHL2) animals (Lin et al., 2001) were obtained from Jackson laboratories and maintained on the congenic C57BL/6 background. To continuously deliver compounds, osmotic pumps delivering 0.25 µL/hr (Model 2004) were used to deliver 300 µg/day of sssiRNA or phosphate buffered saline (PBS) (Sigma Aldrich) for 28 days and pumps designed to deliver 0.5 µL/hr (Model 2002) were used to deliver 75 µg/day of the positive control MOE ASO for 14 days. Pumps (Durect Corporation) and were filled with ss-siRNA or MOE diluted in sterile PBS and then incubated at 37°C for 24 or 48 (2004) hours prior to implantation. Mice were anesthetized with 2.5 % Isofluorane and a midline incision was made at the base of the skull. Using stereotaxic guides, a cannula was implanted into the right lateral ventricle and secured with Loctite adhesive. A catheter attached to an Alzet osmotic mini pump was attached to the cannula and the pump was placed subcutaneously in the midscapular area. The incision was closed with 5.0 nylon sutures. Animals were sacrificed 4 weeks after initiating treatment. Brains were sectioned into 1–2mm coronal sections and frozen on dry ice and stored at −80. Brain regions were harvested for RNA and biochemical analysis using 2 mm punches.
Supplementary Material
Table 1.
| No. | Abbr. Name |
Sequence | Position of Mismatch |
Tm, °C | mut IC50 (nM) |
wt IC50 (nM) | Selectivity (fold) |
|---|---|---|---|---|---|---|---|
| ssRNAs with 5’-(E)-vinyl phosphonate chemistry: | |||||||
| 537787 | REP | None | 90–91 | 8.0 ± 1.7 | > 100 | >13 | |
| 537775 | P9 | 9 | 84–85 | 3.5 ± 0.3 | > 100 | >29 | |
| 537786 | P10 | 10 | 84–85 | 22.3 ± 2.6 | > 100 | >4 | |
| ssRNAs with 5’-phosphate chemistry: | |||||||
| 553819 | REP | None | > 95 | 5.7 ± 1.4 | 71.8 ± 15.6 | 13 | |
| 556886 | P4 | 4 | 85 | 8.8 | 61.1 | 7 | |
| 556887 | P5 | 5 | 82–89 | 17.2 ± 2.6 | >100 | >6 | |
| 556888 | P6 | 6 | 81–85 | N.I. | N.I. | N/A | |
| 556889 | P7 | 7 | 83–85 | 11.2 ± 1.7 | 51.1 ± 8.5 | 5 | |
| 556890 | P8 | 8 | 83–85 | 4.1 ± 0.7 | 29.5 ± 4.3 | 7 | |
| 553822 | P9 | 9 | 85 | 4.9 ± 0.8 | 90.4 ± 9.7 | 18 | |
| 553821 | P10 | 10 | 82–86 | 17.8 ± 3.5 | >100 | >6 | |
| 557407 | P10R | 10 | 77–80 | 15.3 ± 2.4 | >100 | >7 | |
| 556891 | P11 | 11 | 78–81 | 12.8 ± 1.7 | >100 | >8 | |
| 556892 | P12 | 12 | 81–84 | 3.4 ± 0.6 | 72.0 ± 12.3 | 21 | |
| 557406 | P13 | 13 | 78–82 | 4.2 ± 0.6 | >100 | >24 | |
| 557408 | P16 | 16 | 81–84 | 8.1 | 27.3 | 3 | |
| 557409 | P910 | 9, 10 | 78–81 | 6.3 ± 0.5 | >100 | >16 | |
| 557426 | PM3 | 9, 10, 11 | 74 | 3.3 ± 0.5 | >100 | >30 | |
| 557427 | PM4 | 8, 9, 10, 11 | 65–66 | 11.8 ± 1.9 | >100 | >8 | |
| 557428 | RM3 | 4, 10, 16 | 65 | 22.3 | >100 | >4 | |
| 557429 | RM4 | 3, 8, 13, 17 | 65–66 | N.I. | N.I. | N/A | |
| 557430 | REPU | None | >95 | 19.4 ± 4.7 | >100 | >5 | |
| Control oligomers: | |||||||
| 388916 | Gapmer | N/A | N/A | 7.4 ± 0.7 | 12.6 ± 1.3 | 1.7 | |
| 387898 | Gapmer | N/A | N/A | 5.8 | 7.1 | 1.2 | |
| 522247 | Neg.Ctrl | N/A | N/A | N.I. | N.I. | N/A | |
| N/A | LNAT | None | 97 | 40 ± 7 | >100 | >2.5 | |
| N/A | CM |
5’-GCUAUACCAGCGUCGUCAUAA-3’ 3’-TTCGA UAUGGUCGCAGCAGUA-5’ |
N/A | N/A | N.I. | N.I. | N/A |
Color schemes represent different chemical modifications as follows:

All other sugars are riboses unless specifiied otherwise.
N.I.: no inhibition; N/A: not available.
Except for CM (a dsRNA species), all Tm’s are measured using ss-siRNAs duplexed with equi-molar of unmodified ssRNA of the sequence 5’-CAGCAGCAGCAGCAGCAGCAGC-3’.
Gapmers are entirely of phosphorothioate backbone, with 10 central bases having deoxy-ribose and each 5 flanking bases being 2’-MOE chemistry.
All data were obtained in HD-patient-derived fibroblast GM 04281 cell-line. Selectivity is calculated by dividing IC50 of wild-type HTT by that of mutant HTT. Error ranges represent standard error of the mean (SEM) of IC50 values from biological replicates or multiplicates.
Article Highlights.
-
-
Single-stranded RNA functions through RNAi in cells
-
-
Single-stranded RNA achieves allele-selective inhibition of huntingtin
-
-
Allele-selective inhibition proceeds through a miRNA-like mechanism
-
-
Direct ssRNA infusion reduces mutant huntingtin levels in mouse brain.
ACKNOWLEDGEMENTS
Work in the Corey Laboratory was supported by the US National Institutes of Health (NIGMS 73042), an award from the McKnight Foundation for Neuroscience, CHDI Foundation Inc., and the Robert A. Welch Foundation (I-1244). We thank Hui Tian and Haoming Liu of Qinghua Liu’s Lab for generously providing us with recombinant hAgo2 proteins and the training on Ago2-activity assay and Keith Gagnon for helpful comments and for assistance setting up the in vitro AGO2 and RNAse H assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures and four figures.
REFERENCES
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch DA, Jensen S, Liu Y, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson BL, McCray PB. Current prospects for RNA interference based therapies. Nature Rev. Genetics. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mezer M, Wajciechowska M, Naperala M, Sobczak K, Kryzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acid Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, Brouillet E, Luthi-Carter R, Hantraye P, Deglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat. Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Fiszer A, Mykowska A, Krzyzosiak WJ. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S. Huntington’s Disease. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a007476. a007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Pendergraff H, Deleavey G, Swayze E, Potier P, Randolph J, Roesch E, Chattopadhyaya J, Damha M, Bennett F, Chrisophe M, Lemaitre M, Corey DR. Allele-selective silencing of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Watts JK, Pendergraff HM, Montaillier C, Thi D, Potier P, Corey DR. Antisense and antigene inhibition of gene expression by cell-permeable oligonucleotide-oligospermine conjugates. J. Am. Chem. Soc. 2011;133:8404–8407. doi: 10.1021/ja200312y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin JD, Colombo K, Molina—Calavita M, Keryer G, Zala D, Charrin BC, Dietrich P, Vovert M-L, Guillemot F, Dragatsis I, et al. Huntingtin is required for Mitotic Spindle Orientation and Mammalian Neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- Hall AHS, Wan J, Spesock A, Sergueeva Z, Ramsay Shaw B, Alexander KA. High potency silencing by single-stranded boranophosphate siRNA. Nucleic Acids Res. 2006;34:2773–2781. doi: 10.1093/nar/gkl339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigsma HJ, Li JJ, Soriano F, Kenski DM, Flanagan WM, Willingham AT. mRNA knockdown by single-stranded RNA is improved by chemical modifications. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1301. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Babaie E, Prydz H. Similar behavious of single and double strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009c;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Gagnon KT, Liu J, Watts JK, Syeda-Nawaz J, Bennett CF, Swayze EE, Randolph J, Chattopadhyaya J, Corey DR. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol. Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Corey DR. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Sanders SS, Kang R, Carroll JB, Sutton L, Wan J, Roshini S, Young FB, Liu L, El-Hesseini A, et al. Wild-type HTT modulates the enzymatic activity of the neuronal palmitoyl transferase HIP14. Hum. Mol. Genet. 2011;20:3356–3365. doi: 10.1093/hmg/ddr242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, Seth PP, Swayze EE, Crooke ST. Single-stranded siRNAs activate RNAi in animals. Cell. 2012 doi: 10.1016/j.cell.2012.08.014. in Press. [DOI] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, Artates JW, Cheng SH, Shihabuddin LS, Hung G, Bennett CF, Cleveland DW. Sustained therapeutic reversal of Huntington’s disease by transient repression of mutant huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski P, de Mezer, Krzyzosiak WJ. Trinucleotide repeats in the human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer B, Goldberg P, Andrew SE, Theilmann J, Telenius H, Zeisler J, Squitieri F, Lin B, Bassett A, Almqvist E, et al. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. N. Engl. J. Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E, Donato SD, Kaemmerer WF. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matsui M, Corey DR. Drug Discov. Today. 2012;17:43–450. doi: 10.1016/j.drudis.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Omi K, Hachiya NS, Tokunaga K, Kaneko K. siRNA-mediated inhibition of endogenous Huntington disease gene expression induces an aberrant configuration of the ER network in vitro. Biochem. Biophys. Res. Commun. 2005;338:1229–1235. doi: 10.1016/j.bbrc.2005.10.061. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah DWY, Aronin N. Oligonucleotide therapeutic approaches for Huntingtin Disease. J. Clin. Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum. Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, Collins JA, Semaka A, Hudson TJ, Hayden MR. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 1997;17:404–410. [Google Scholar]
- Zeitlin S, Liu J-P, Chapman DL, Papaioannnou VE, Estratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.