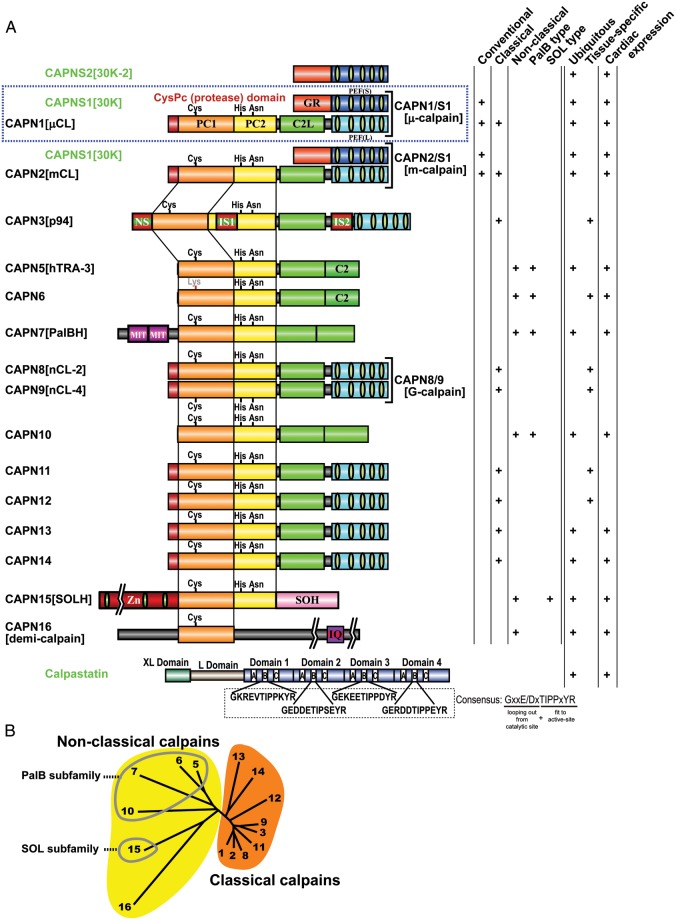
Figure 2.
Human calpains and their regulatory molecules. (A) Schematic structures. In this review, human calpains are presented in a proposed name (previous name, if any) format. Calpain structural classification and tissue distribution, particularly regarding the heart, is indicated in the right-hand columns. Calpain enzyme complexes (tertiary structures) that have been elucidated in vivo are shown along with their enzyme names (single brackets). Bottom: the domain structure of the longest calpastatin isoform. The four repetitive inhibitory units are labelled as domains 1–4; in each of these, the A–C regions are important for calpastatin's inhibitory activity. The consensus aa sequence in the B-region, which directly interacts with the calpain active site, is shown. Exons encoding the XL and L domains are subject to alternative splicing. Symbols: PC1 and PC2, protease core domains 1 and 2 in the calpain protease (CysPc) domain; C2L, C2-domain-like domain; PEF(L) and PEF(S), penta-EF-hand domains in the larger (L) and smaller (S) subunits, respectively; GR, glycine-rich hydrophobic domain; NS/IS1/IS2, CAPN3[p94]-characteristic sequences; MIT, microtubule interacting and trafficking motif; C2, C2 domain; Zn, Zn-finger motif domain; SOH, SOL-homology domain; IQ, a motif that interacts with calmodulin. (B) Human calpain (represented by its number) phylogenetic tree, drawn using the neighbour-joining and bootstrap method after aligning all sequences.4 Non-classical calpains are divided into further subfamilies.