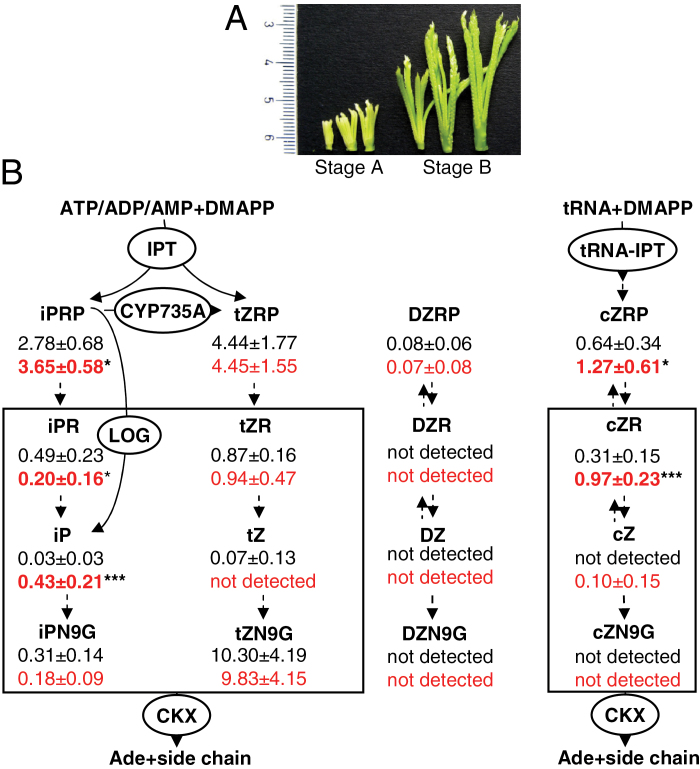
Fig. 3.
(A) General view of developing finger millet inflorescences used for cytokinin measurements as well as for molecular biological analyses. (B) Levels of cytokinins measured in young inflorescences (stage A) of SE7 and wild-type finger millet. Wild-type cytokinin contents are depicted in black; and those of the SE7 mutant in red. Data represent mean values (in pmol g–1 fresh weight) ±SD. Values representing significant differences between SE7 and the wild type are shown in bold (*P < 0.05; ***P < 0.001, calculated by Student’s t-test). Key enzymes involved in cytokinin biosynthesis and degradation are shown in circles. Cytokinin derivatives shown in a box are potential targets for the CKX enzyme (Frébort et al., 2011). CKX, cytokinin oxidase/dehydrogenase; cZ, cis-zeatin; cZNG, cis-zeatin 9-glucoside; cZR, cis-zeatin riboside; cZRP, cis-zeatin ribotide-phosphate; DZ, dihydro-zeatin; DZNG, dihydro-zeatin 9-glucoside; DZR, dihydro-zeatin riboside; DZRP, dihydro-zeatin ribotide-phosphate; iP, N 6-(Δ2-isopentenyl) adenine; iPR, iP riboside; iPNG, iP 9-glucoside; iPRP, iP ribotide-phosphate; IPT, adenosine phosphate-isopentenyltransferase; LOG, LONELY GUY; tRNA-IPT, tRNA isopentenyltransferase; tZ, trans-zeatin; tZNG, trans-zeatin 9-glucoside; tZR; trans-zeatin riboside; tZRP, trans-zeatin ribotide-phosphate.