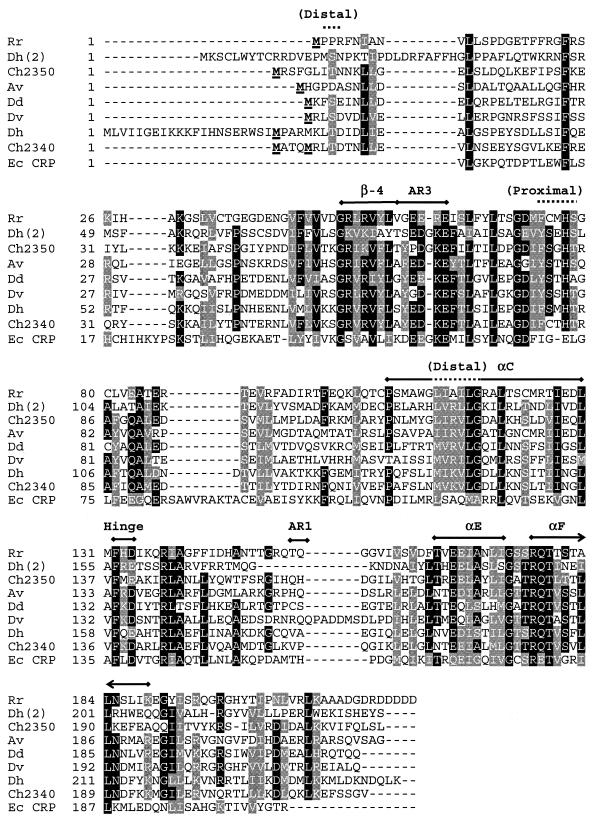
FIG. 2.
Sequence alignment of CooA homologs. Eight CooA homologs and CRP from E. coli were aligned by using the T-Coffee multiple sequence alignment tool (version 1.37) as implemented at http://www.cmbi.kun.nl/bioinf/tools/T_COFFEE/ (25). Putative CooA homologs were identified by a TBLASTN search of 207 completed and unfinished microbial genomes available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) queried with the R. rubrum CooA protein. Designations of the aligned sequencesand their accession parameters are as follows: Rr, R. rubrum (gi no. 1498752); Dh(2), D. hafniense (gi no. 23114031); Ch2350, C. hydrogenoformans (gnl no. TIGR_129958, contig 2350:c_hydrogenoformans); Av, A. vinelandii (gi no. 23105899); Dd, D. desulfuricans G20 (gi no. 23473780); Dv, D. vulgaris (gnl no. TIGR_881, 1531); Dh, D. hafniense (gi no. 23111778); Ch2340, C. hydrogenoformans (gnl no. TIGR_129958, contig 2340:c_hydrogenoformans); Ec CRP, E. coli (gi no. 117484). Shaded portions indicate 70% conservation, with key structural elements indicated above sections of the alignment. These include residues that form the distal and proximal heme ligand environments (overlined with a dotted line) as well as conserved structural elements including the His heme ligand (His77 in R. rubrum CooA), the β-4 sheet, helices αC, αE, and αF, the hinge sequence between the effector and DNA-binding domains, and the AR1 and AR3 regions that interact with RNA polymerase (overlined with a thick line). Boldface underlined residues at or near the N terminus represent the cloned initiation of the proteins; for the CooA from R. rubrum, the terminal Met is removed posttranscriptionally.