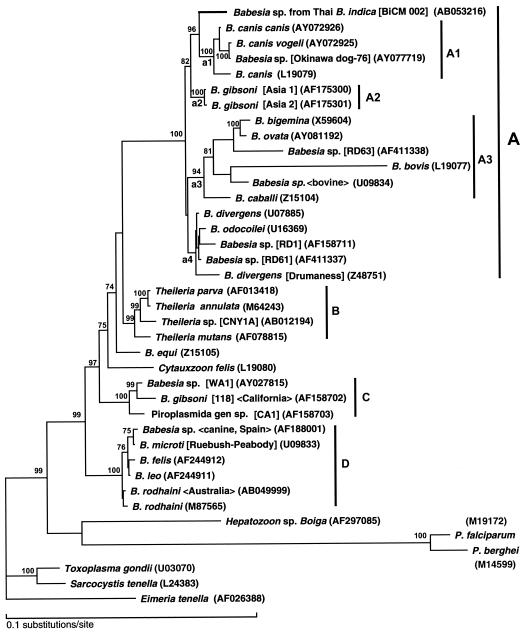
FIG. 2.
Phylogenetic tree of the SSUrDNA sequences of piroplasms. The best tree finally selected by the maximum-likelihood analysis is shown. Details of the phylogenetic analysis will be described elsewhere (A. Dantrakool, T. Hashimoto, A. Saito et al., unpublished data). Briefly, based on the result obtained by use of FASTA, in total 33 SSUrDNA sequences from Babesia spp., Theileria spp., Cytauxzoon spp., and unclassified piroplasms were included in the data set for the analysis, including the sequence of the Babesia spp. from Thai Bandicota indica. Two SSUrDNA sequences of Plasmodium spp. and the sequence of Hepatozoon sp. Boiga were added to the data set. Three sequences of other apicomplexan parasites, Toxoplasma gondii, Sarcocystis tenella, and Eimeria tenella, were also included as outgroups, and 1,378 unambiguously aligned positions were selected and used. Bootstrap proportions (percent) with more than 70% support are attached to the internal branches. Monophyletic groups and subgroups supported with more than 90% in the analysis are shown by vertical bars. The horizontal length of each branch is proportional to the estimated number of substitutions. GenBank accession numbers are given in parentheses. The name of the isolate, strain, clone or genotype is shown in brackets. The host origin or the isolation site is given within chevrons (<>).