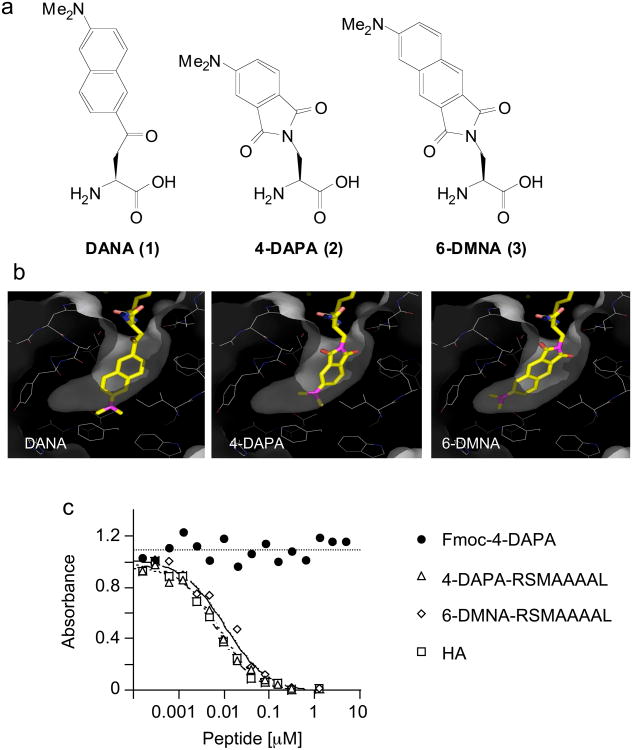
Figure 1. 4-DAPA and 6-DMNA can be modeled into the P1 pocket of DR1 without major distortion.
(a) Chemical structures of DANA, 4-DAPA and 6-DMNA. (b) In the crystal structure of the DR1-HA peptide complex, a tyrosine residue is buried deep into the hydrophobic P1 pocket 5. DANA, 4-DAPA and 6-DMNA were modeled in silico into this pocket in place of tyrosine. Residues shown lining the pocket are (clockwise from upper right) Pheα54, Pheα32, Trpα43 (behind), Ileα7, Trpβ153, Pheα48, Thrβ90, Valβ91, Tyrβ83, Glyβ86, and Valβ85. The side chains of Asnβ82 and Hisβ81 (upper left) form hydrogen bonds with the peptide main chain at the mouth of the P1 pocket. Other residues lining the pocket but not shown are Pheα24, Pheα26, and Pheα48 (c) Binding of fluorogenic peptides to DR1 assessed using a competitive binding assay. DR1 was incubated with biotin-HA peptide and various concentrations of unlabelled inhibitor peptides, with biotin-HA binding quantified by a sandwich ELISA assay using streptavidin alkaline phosphatase. Binding of Fmoc-(4-DAPA) was assessed to evaluate non-specific binding of the fluorophore. IC50 values for these and other peptides (Supplementary Table I online) were determined as described (Supplementary Methods online).