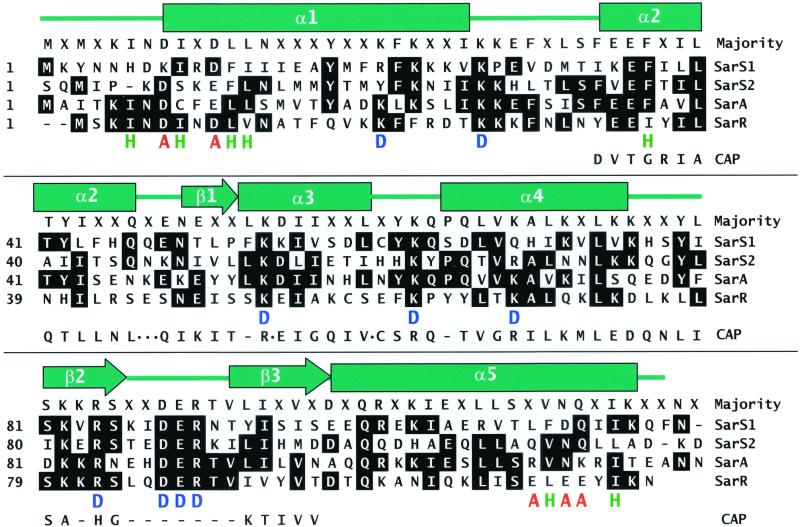
Figure 3.
A sequence alignment of SarA homologs from S. aureus. The sequences are presented in one-letter amino acid code. Numbers at the beginning of each line indicate amino acid positions relative to the start of each protein sequence. Helices are indicated by rectangles, β-sheets are indicated by arrows, and loops are indicated by a line. “H” marked in green represents a residue that takes part in dimerization. “A” marked in red represents a residue that may compose of the activation motifs. “D” marked in blue represents a residue that may be involved in the interaction of SarR with DNA. The sequence of DNA binding motif from CAP is also aligned to the SarA proteins. A dot represents a residue(s) omitted and a dash represents a residue(s) missed in CAP. SarS2 starts at 1 (actual position on SarS is 126).