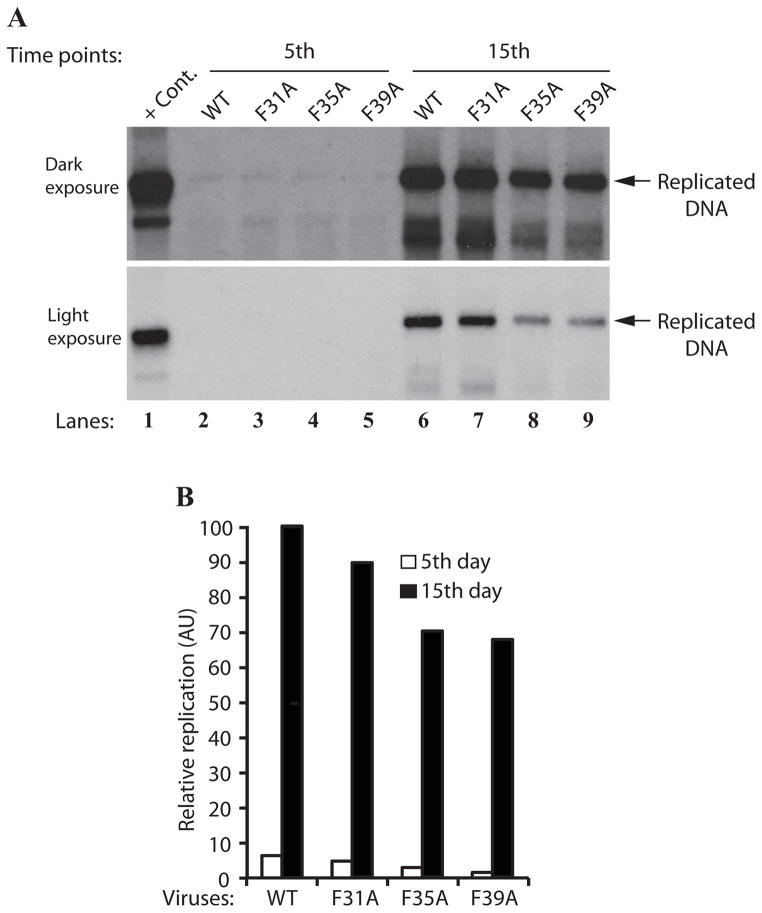
Figure 6.
Effect of Phe mutations on the JCV DNA replication. (A) DpnI assay. SVG-A cells (2×106 cells/75cm2 flask) were separately transfected/infected with JCV Mad-1 WT, JCV Mad-1 Agno-F31A, JCV Mad-1 Agno-F35A or JCV Mad-1 Agno-F39A viral DNA genomes (8 μg each). The low-molecular-weight DNA containing both input and replicated viral DNA was isolated using Qiagen spin columns (Ziegler et al., 2004), and digested with BamHI and DpnI restriction enzymes. Digested DNA was separated on a 1% agarose gel, transferred onto a nitrocellulose membrane (Bio-Rad) and probed for detection of the newly replicated DNA using a probe prepared from the JCV Mad-1 WT as a template. In lane 1, 2 ng of JCV Mad-1 WT linearized by BamHI digestion was loaded as positive control (+ Cont.). Replication assays were repeated several times and a representative result is shown here. (B) Quantitation analysis of Southern blots by a semi-quantitative densitometry method (using NIH Image J program) and presentation of the results in arbitrary units. The replication efficiency of each data point was presented relative to that of WT.