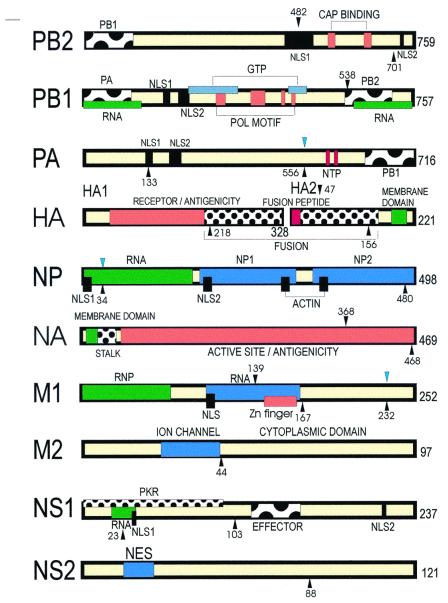
Figure 4.
Location of mutations selected on mouse adaptation on the functional genetic map of Influenza A virus. The location of mutations seen in FM-MA that have been confirmed to control virulence (9) are shown with black downward arrows, and those preexisting in FM virus are indicated in blue. Mutations observed in HKMA clones are shown with upward arrows. The functional regions and binding sites are described elsewhere (12). NLS, nuclear localization signal; POL, polymerase; NTP, nucleotide triphosphate; PKR, dsRNA-dependent protein kinase; effector indicates a multiple protein-binding region; NES, nuclear export signal; regions labeled with influenza virus protein names indicate sites of interaction.