Understanding the molecular pathways and physiological processes underlying aging has universal importance. The p53 family member TAp73, which contains a transcriptional transactivation domain, has been known to play a role in cancer. Here, Rufini et al. show that TAp73 plays a protective role in aging in both human and mouse cells by regulating the expression of Cox4i1, a subunit of the COX multimeric protein complex, thereby affecting mitochondrial respiration as well as preventing the accumulation of reactive oxygen species.
Keywords: p73, p53, aging, senescence, metabolism, ROS, mitochondria
Abstract
Aging is associated with impaired scavenging of reactive oxygen species (ROS). Here, we show that TAp73, a p53 family member, protects against aging by regulating mitochondrial activity and preventing ROS accumulation. TAp73-null mice show more pronounced aging with increased oxidative damage and senescence. TAp73 deletion reduces cellular ATP levels, oxygen consumption, and mitochondrial complex IV activity, with increased ROS production and oxidative stress sensitivity. We show that the mitochondrial complex IV subunit cytochrome C oxidase subunit 4 (Cox4i1) is a direct TAp73 target and that Cox4i1 knockdown phenocopies the cellular senescence of TAp73-null cells. Results indicate that TAp73 affects mitochondrial respiration and ROS homeostasis, thus regulating aging.
Aging is a progressive functional decline that compromises fitness and predisposes to diseases such as cancer and neurodegeneration. Several mechanisms impact on longevity, such as altered insulin/IGF signaling, stem cell depletion, and increased oxidative damage due to impaired scavenging of reactive oxygen species (ROS) (Kenyon 2010). In addition to the mammalian target of rapamycin (mTOR) and the AMP-activated protein kinase (AMPK) regulator of forkhead box O (FOXO), the metabolic pathways of aging involve the AMPK-activated PPARγ coactivator 1α (PGC1α), a regulator of mitochondrial function (Sahin and Depinho 2012). Indeed, reduced mitochondrial bioenergetics, together with increased ROS production, is associated with aging (Balaban et al. 2005), suggesting that progressive mitochondrial dysfunction plays an important role in the aging process.
p73 and p63 have been identified as the ancestral members of the p53 family (Belyi et al. 2010); hence, p73, being a structural and functional homolog of the tumor-suppressing transcription factor p53, can transactivate p53-responsive genes and mediate cell cycle arrest and apoptosis in response to DNA damage (Melino et al. 2002; Levine et al. 2011). Despite the high sequence and structural similarity, the mouse models revealed a crucial role in neural development for p73 and in epidermal formation for p63, unlike the tumor susceptibility of p53 knockout mice. The Tp73 gene is expressed via two distinct promoters as proteins with (TAp73) or lacking (ΔNp73) the transactivation domain (Melino et al. 2002). Although implicated in the regulation of tumorigenesis and in the chemotherapeutic response (Melino et al. 2002), p73 is essential for proper development of the CNS (Yang et al. 2000; Tomasini et al. 2008; Wilhelm et al. 2010). Indeed, selective TAp73 knockout mice develop tumors and show hippocampal dysgenesis (Tomasini et al. 2008), while ΔNp73 knockout mice do not develop tumors but develop late signs of moderate neurodegeneration (Wilhelm et al. 2010).
Here, we report the ability of TAp73 to regulate mitochondrial respiration by directly transactivating the mitochondrial complex IV subunit cytochrome C oxidase subunit 4 (Cox4i1). Reduction of Cox4i1 in TAp73-deficient mice results in premature aging secondary to mitochondrial dysfunction and increased ROS generation, and complementation of TAp73 knockout cells with Cox4i1 rescues the reduced mitochondrial oxygen consumption.
Results and Discussion
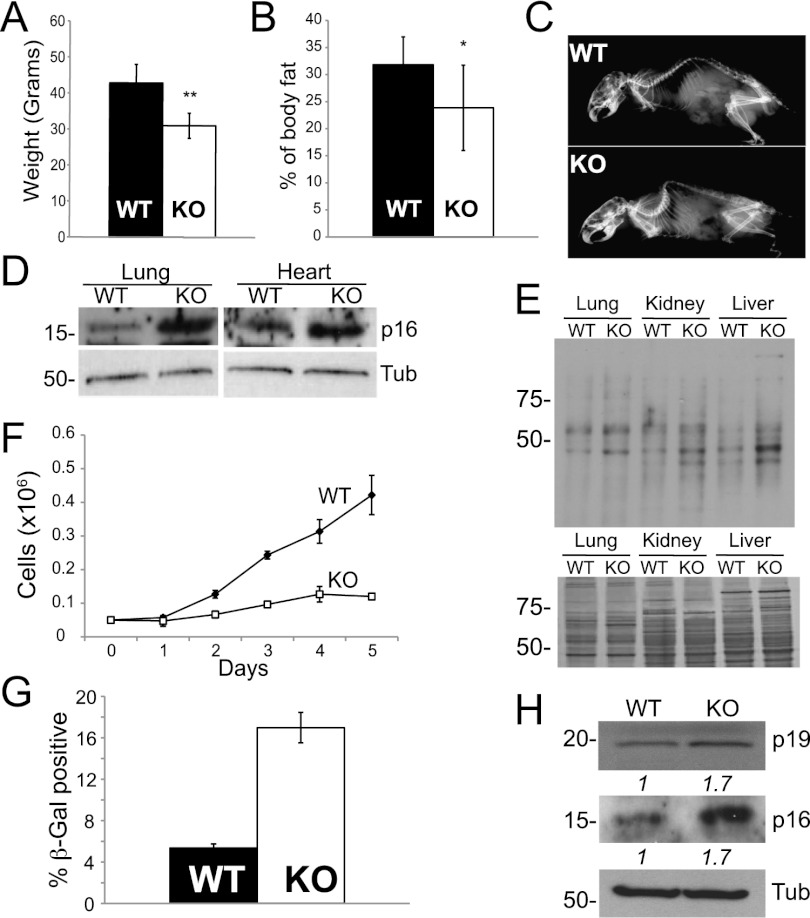
We previously established that TAp73 deficiency predisposes mice to tumorigenesis (Tomasini et al. 2008). To assess whether loss of TAp73 also influenced the aging process, we studied a number of age-related characteristics in old tumor-free animals. First, TAp73 knockout mice showed reduced survival (mean 19.1 ± 1.2 mo, 50% survival [n = 22], compared with 25.0 ± 2.1 mo in wild-type mice [n = 18]; P < 0.01) (see also Tomasini et al. 2008). In addition, 18-mo-old TAp73 knockout mice had lower body weights with a decreased percentage of body fat, particularly subcutaneous fat (Fig. 1A,B; Supplemental Fig. 1A) compared with their wild-type controls. The knockout mice also had radiological evidence of severe kyphosis (Fig. 1C), with marked thinning of the epidermis (Supplemental Fig. 1B) and corneal degeneration (Supplemental Fig. 1C). In addition, when subjected to a hair growth assay, all of the TAp73-null animals showed essentially no hair growth 20 d after shaving, compared with robust growth in wild-type mice (Supplemental Fig. 1D). IGF1 serum levels decrease during aging, and TAp73 knockout mice also had reduced serum levels of IGF-1 (Supplemental Fig. 1E). Western blot analysis of several organs revealed elevated levels of p16, a well-established biomarker of aging (Fig. 1D; Krishnamurthy et al. 2004; data not shown). Finally, immunoblotting of carbonyl groups showed increased levels of oxidized proteins in the livers, kidneys, and lungs from knockout mice (Fig. 1E), suggesting increased oxidative stress in vivo. Together, these data indicate that TAp73 deficiency aggravates age-related features and increases expression of markers of oxidative stress and senescence in vivo.
Figure 1.
TAp73 depletion accelerates aging and senescence. (A) Reduced body weight affecting TAp73 knockout (KO) animals (n = 8). Mean ± SD. (B) Percentage of body fat measured with Lunar PIXImus densitometry (n = 8). Mean ± SD. (C) Radiography showing enhanced kyphosis in 18-mo-old TAp73 knockout mice. (D) p16 levels detected by Western blot in organ extracts from old animals. Tubulin was used as loading control. (E, top) Levels of carbonylated proteins in the indicated organs of 18-mo-old wild-type (WT) and TAp73 knockout mice. (Bottom) Coomassie staining of organ extracts shows even loading. (F) Decreased proliferative capacity in late passage (P8) TAp73 knockout MEFs versus wild-type control. (G) Quantification of SA-β-Gal-positive senescent MEFs. Mean ± SD. (H) Western blot of senescence markers p16 and p19ARF in late passage (P8) MEFs. Tubulin was used as loading control.
To investigate the mechanism responsible for the accelerated aging in TAp73 knockout mice, we generated mouse embryonic fibroblasts (MEFs) from embryonic day 14.5 (E14.5) null and control embryos, passaged them in vitro according to the standard 3T3 protocol, and assessed their propensity to undergo replicative senescence in late passages (P7–P8). We found that, unlike their wild-type counterparts, late passage TAp73 knockout MEFs ceased to proliferate (Fig. 1F) and showed increased β-galactosidase activity (SA-β-gal) (Fig. 1G), suggesting that TAp73 depletion triggers senescence in vitro. As senescent cells also display increased expression of p19ARF and p16, we investigated the expression of these markers. Consistently, late passage TAp73 knockout MEFs expressed increased levels of p19ARF and p16 proteins with respect to wild-type cells (Fig. 1H). Overall, these data indicate that loss of TAp73 accelerates senescence in vitro, supporting the age-related symptoms observed in the knockout animals.
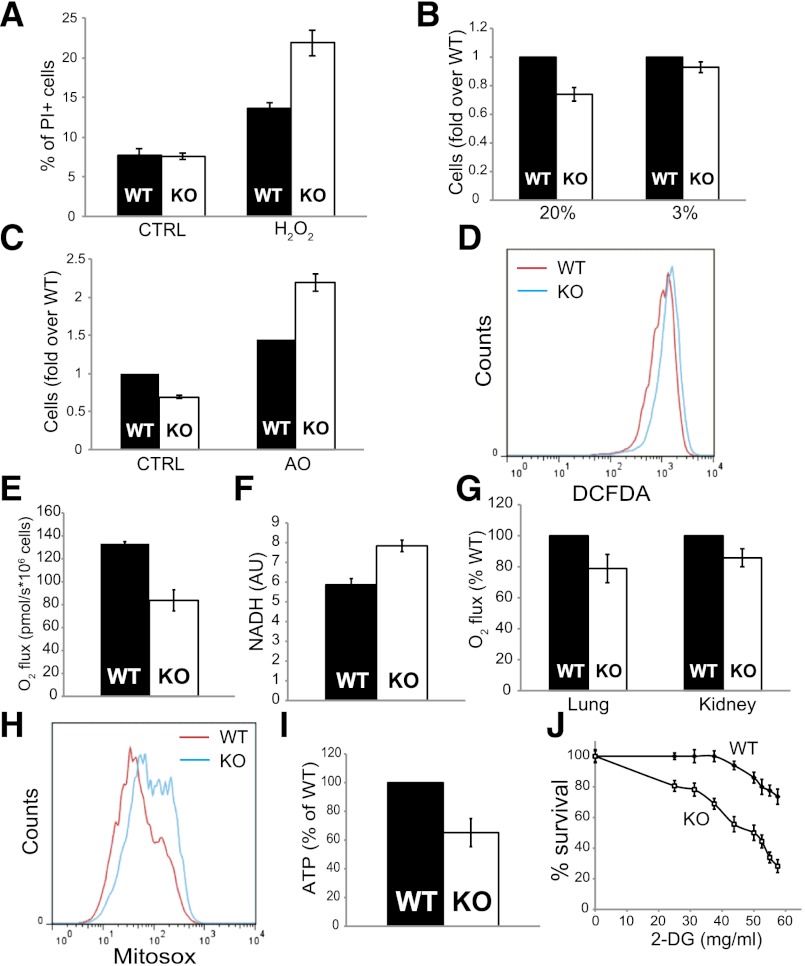
Previous studies showed that senescence in mouse fibroblasts is the outcome of sustained oxidative damage (Parrinello et al. 2003) in standard culturing conditions. Indeed, MEFs accumulate oxidative damage when cultured in atmospheric (i.e., 20%) oxygen. This “oxygen sensitivity” slows cell proliferation and triggers senescence, which can be reverted by switching growth conditions to 3% oxygen (Parrinello et al. 2003). Therefore, we reasoned that deletion of TAp73 might sensitize cells to oxidative damage. To test this possibility, we challenged early passage (P2), presenescent wild-type and knockout MEFs with 0.5 mM hydrogen peroxide (H2O2) for 16 h and assayed cell death by propidium iodide (PI) exclusion. Notably, TAp73 knockout MEFs were more sensitive than their wild-type counterparts to H2O2-induced death, with an ∼50% increase in cell death (Fig. 2A). Next, we investigated the effect of oxygen concentration on the proliferative capacity of early passage MEFs (P2). After 6 d in culture in a 20% oxygen atmosphere, there were fewer TAp73 knockout cells, although knockout and wild-type cells proliferated equally when cultured in low oxygen (3%) (Fig. 2B). A similar rescue of the growth retardation of TAp73 knockout MEFs was achieved by addition of antioxidants (Fig. 2C). Of note, antioxidants were also able to revert the accumulation of p16 and SA-β-Gal in late passage MEFs from both wild-type and TAp73 knockout mice (Supplemental Fig. 2), further supporting the notion that oxidative damage elicits senescence in mouse fibroblasts. Overall, these data suggest that TAp73 protects against oxidative damage and that its depletion slows cell proliferation and elicits senescence consequent to increased susceptibility to oxygen-associated damage.
Figure 2.
Reduced mitochondrial function and unbalanced ROS in TAp73 knockout early passage MEFs. (A) Wild-type (WT) and TAp73 knockout (KO) MEFs (p2) were left untreated (CTRL) or challenged with 0.5 mM H2O2 (H2O2), and cell death was measured by PI staining and flow cytometry. Mean ± SD. (B) Wild-type and TAp73 knockout MEF growth at 20% and 3% oxygen concentration. Cell number is expressed as fold over wild-type control. Mean ± SD. (C) Wild-type and TAp73 knockout MEFs were treated with vehicle (CTRL) or 30 μM Trolox/30 μM ascorbic acid antioxidant mixture (AO). Cells were allowed to grow for 72 h before counting. Cell number is expressed as fold over wild-type control. Mean ± SD. (D) Increased staining of TAp73 knockout MEFs with the ROS fluorophore DCFDA. (E) State III oxygen consumption in wild-type and TAp73 knockout MEFs. Oxygen consumption was measured with an Oroboros oxygraph and normalized to cell number. State III was induced by addition of ADP. (F) NADH levels measured in MEFs and normalized to protein content. Mean ± SD. (G) Reduced oxygen consumption in the lungs and kidneys from wild-type and TAp73 knockout mice. Data are expressed as percentage of wild-type controls (n = 3). (H) Measurement of mitochondrial O2− levels with MitoSox Red in wild-type and knockout MEFs. (I) ATP levels in MEFs normalized to protein content and expressed as percentage of wild-type control. (J) Wild-type and knockout MEFs treated with the indicated doses of 2-deoxy-D-glucose (2-DG) were assayed for survival by trypan blue exclusion. Mean ± SD.
Next, we sought to investigate whether the oxidative imbalance affecting TAp73 knockout cells was accompanied by altered levels of intracellular ROS. To this end, we measured ROS content in early passage MEFs (P2) with the radical dye dichlorofluorescein diacetate (DCFDA). Importantly, the susceptibility of TAp73-depleted cells to oxidative damage correlated with a robust increase in intracellular ROS (Fig. 2D).
In an attempt to understand the reason for this increase, we focused on mitochondria, since mitochondria have been shown to play a fundamental role in oxidative metabolism and aging (Sahin and Depinho 2012) and represent the main cellular source of ROS. Moreover, p53 is known to regulate mitochondrial oxygen consumption through multiple pathways (Gottlieb and Vousden 2010). Since p73 is part of the p53 superfamily, we asked whether the increased ROS levels triggered by depletion of TAp73 depend on altered mitochondrial activity. We assessed aerobic respiration in TAp73 knockout and wild-type MEFs and found that TAp73-depleted cells had significantly decreased rates of both basal and state III (i.e., ADP-driven) oxygen consumption (Fig. 2E; Supplemental Fig. 3A). Direct measurement of mitochondrial electron transport chain (ETC) enzymatic activity showed a consistent selective reduction in complex IV cytochrome C oxidase (COX) activity (Supplemental Fig. 3B). This reduced flow of electrons through the ETC would be expected to increase the amount of reduced NADH, the major ETC electron donor. Consistently, NADH levels were increased in TAp73 knockout MEFs when compared with wild-type cells (Fig. 2F). Finally, to assess whether TAp73 depletion affects mitochondrial respiration in vivo, we measured oxygen consumption in the lungs and kidneys from TAp73 knockout mice (which, in wild-type mice, express relatively high TAp73 mRNA levels) and found that oxygen fluxes were consistently decreased (Fig. 2G).
Reduced complex IV activity will retain upstream complexes in a reduced state, thereby favoring electron leakage and increasing mitochondrial ROS production (Liu et al. 2009). To test this possibility, we measured anion superoxide (O2−) in MEFs using the mitochondrial superoxide dye MitoSox Red and found it robustly increased in TAp73 knockout cells (Fig. 2H), suggesting that mitochondrial dysfunction is indeed the major source of the increased ROS levels observed in the absence of TAp73. Finally, we investigated the impact of TAp73 depletion on cellular energetics. TAp73 knockout cells have reduced basal ATP levels compared with parental cells (Fig. 2I), further suggestive of impaired oxidative phosphorylation (OXPHOS). The reduction of OXPHOS contribution to energetic balance is underlined by the increased glucose addiction in TAp73 knockout MEFs, as revealed by their increased sensitivity to glycolysis inhibition by 2-deoxyglucose (Fig. 2J).
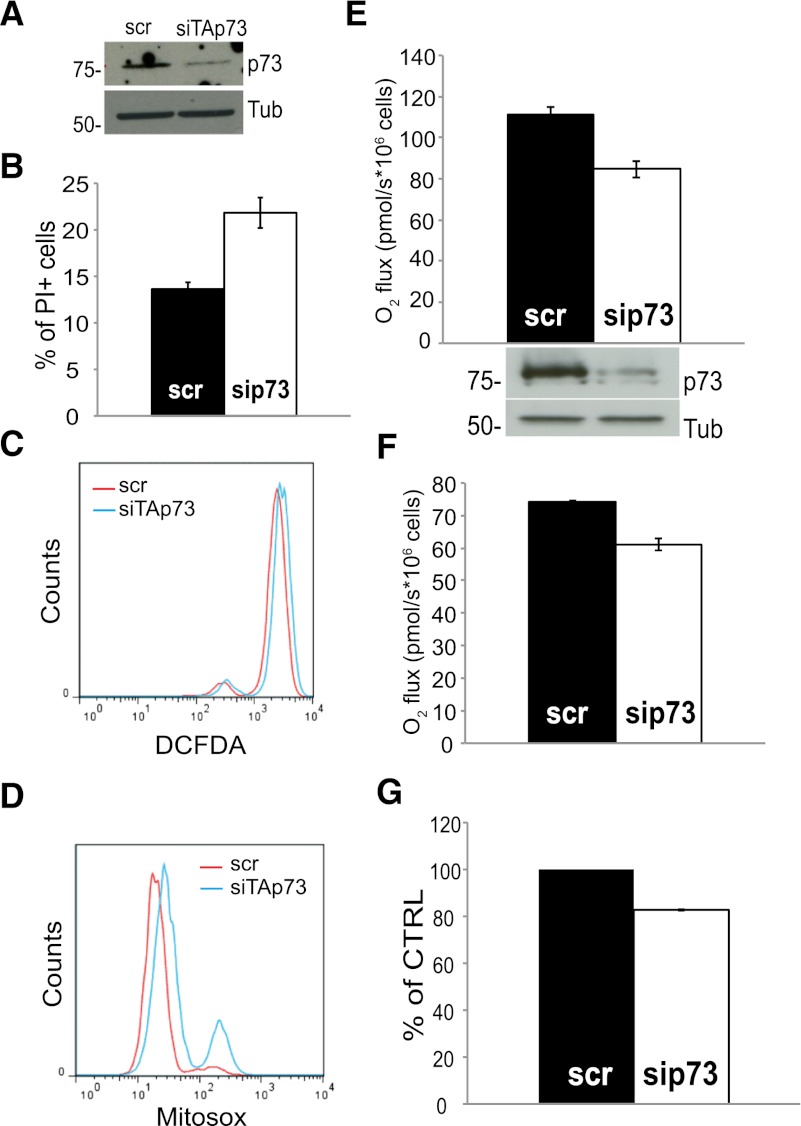
Next, we investigated whether these findings could be extended to human cells. Accordingly, we knocked down TAp73 in the human colon carcinoma HCT116 cell line using siRNA technology to selectively target TAp73 isoforms (Fig. 3A; Supplemental Fig. 4). Like mouse fibroblasts, TAp73-depleted human cells show increased cell death upon H2O2 treatment (Fig. 3B). In addition, knockdown of TAp73 increased intracellular ROS (Fig. 3C) and, more specifically, mitochondrial O2− levels (Fig. 3D). To understand whether TAp73 sustains mitochondrial function in HCT116 cells, we measured oxygen consumption in control and TAp73-depleted cells. Notably, reduced expression of TAp73 results in reduced state III and basal respiratory rates (Fig. 3E,F), accompanied by decreased activity of mitochondrial complex IV (Fig. 3G). Together, these data demonstrate that p73 deficiency compromises cellular bioenergetics and mitochondrial function, resulting in an increased intracellular ROS content.
Figure 3.
Knockdown of TAp73 in HCT116 cells recapitulates the mouse phenotype. (A) Western blot analysis showing the efficiency of TAp73 knockdown. Cells were treated with siRNA control (scr) and siRNA against TAp73 (siTAp73) and assessed at 72 h post-transfection. (B) Cells treated with siRNA control (scr) and siRNA against TAp73 (sip73) were left untreated (CTRL) or challenged with 0.2 mM H2O2 for 8 h (H2O2), and cell death was quantified by PI staining and flow cytometry. Mean ± SD. (C) Analysis of siRNA-treated cells using DCFDA. TAp73-depleted cells show increased staining. (D) Analysis of mitochondrial O2− levels with the selective dye MitoSox Red in TAp73-depleted HCT116. (E) State III oxygen consumption in cells treated with siRNA control or siRNA targeting TAp73. Oxygen consumption was measured with an Oroboros oxygraph and normalized to cell number. State III was induced by injection of ADP. Mean ± SD. (F) Basal respiration of cells treated as in E. Mean ± SD. (G) Activity of the complex IV of the ETC in cells treated with siRNA control or siRNA targeting TAp73.
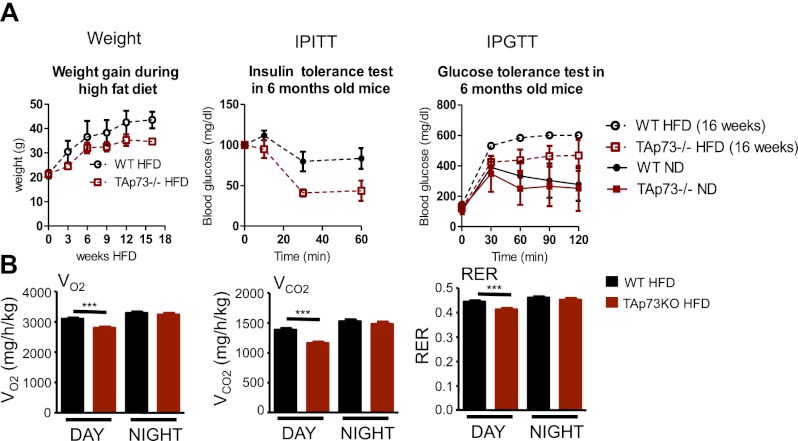
Mice lacking glutathione peroxidase (GPX-1), which therefore cannot detoxify ROS, have been reported to be protected from the insulin resistance induced by a high-fat diet (HFD) (Loh et al. 2009). To test whether TAp73 knockout mice show a similar phenotype, we fed knockout and wild-type littermates with chow (normal diet [ND]) or a HFD (60% kcal from fat). Intraperitoneal glucose tolerance tests (IPGTTs) and intraperitoneal insulin tolerance tests (IPITTs) after 16 wk of the HFD revealed that TAp73 knockout mice were partially protected from glucose intolerance and insulin resistance (Fig. 4A). Analysis of energy expenditure under the same conditions revealed that knockout mice had significantly decreased oxygen consumption (Fig. 4B).
Figure 4.
Effects of TAp73 deficiency on glucose tolerance during high caloric intake. (A) TAp73 knockout (KO) mice showed decreased weight gain and improved insulin sensitivity and glucose tolerance after a 16-wk treatment with a HFD, compared with a ND. (B) TAp73 knockout mice showed decreased VO2, VCO2, and respiratory exchange ratio (RER) after a 16-wk treatment with a HFD. (n = 6 per group). Mean ± SD. (**) P < 0.01.
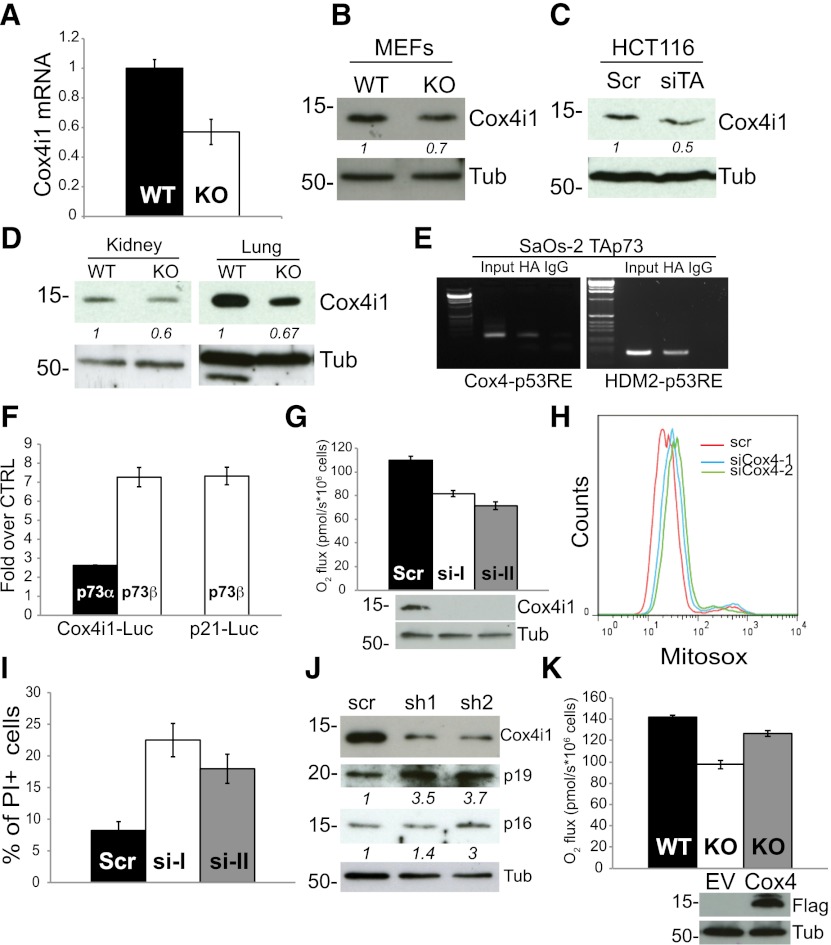
Next, we sought to investigate the molecular mechanism underlying TAp73-mediated regulation of mitochondrial function. We did not observe any differences in mitochondrial structure and mass or find differences in expression of the p53 target PGC1α between wild-type and TAp73 knockout MEFs (Supplemental Fig. 5A–D). However, the decreased enzymatic activity of COX suggests that p73 may regulate expression of genes involved in complex IV function. Recent studies using gene expression profiling have suggested that the p53 family may affect expression of SCO2 and COX4i1 (Matoba et al. 2006; Lin et al. 2009). Complex IV of the ETC contains four main core subunits: Mitochondrial-encoded subunits I, II, and III provide the catalytic core of COX, while the nuclear-encoded subunit IV regulates kinetic properties of the holoenzyme. SCO2 encodes a regulatory protein that delivers copper to the catalytic site of subunit II, while Cox4i1 specifies isoform 1 of the subunit IV of COX. We failed to detect changes in SCO2 transcript levels in TAp73 knockout cells (Supplemental Fig. 5E). In contrast, TAp73 knockout MEFs expressed reduced mRNA and protein levels of Cox4i1 (Fig. 5A,B). Cox4i1 protein content was also reduced after siRNA-mediated knockdown of TAp73 in HCT116 cells (Fig. 5C) and in kidneys and lungs isolated from TAp73 knockout animals (Fig. 5D; Supplemental Fig. 6A). Moreover, ectopic expression of p73 increased Cox4i1 levels in HCT116 (Supplemental Fig. 6B) and in HA-tagged TAp73 SaOs-2–Tet-on cell lines (Supplemental Fig. 6C). These findings and the identification of a p53-responsive element (p53RE) in intron 1 of the mouse and human Cox4i1 locus (see the Supplemental Material) suggest that Cox4i1 may be a p73 target gene. A chromatin immunoprecipitation (ChIP) experiment in SaOs-2–Tet-on-inducible cells showed direct binding of TAp73 to the COX4i1 intronic responsive element (Fig. 5E). We also cloned a 400-base-pair (bp) fragment of the COX4i1 intron 1 containing the p53RE upstream of a luciferase reporter vector and demonstrated that luciferase activity was increased by ectopic expression of TAp73 isoforms (Fig. 5F).
Figure 5.
Cox4i1 is a TAp73 target gene. (A) Quantitative PCR of Cox4i1 transcripts in wild-type (WT) and TAp73 knockout (KO) MEFs normalized to endogenous actin. (B) Protein levels of Cox4i1 in the same cells. Tubulin was used as a loading control. (C) Cox4i1 protein in HCT116 cells treated with siRNA control or siRNA targeting TAp73. (D) Reduced Cox4i1 protein in extracts from the lungs and kidneys of 6-mo-old wild-type and TAp73 knockout mice. (E) ChIP performed on HA-tagged TAp73 Tet-inducible SaOs-2 showing direct binding of TAp73 to the p53RE in the first intron of Cox4i1. HDM2 was used as positive control. (F) Luciferase assay showing TAp73-meditated up-regulation of luciferase activity driven by a 400-bp first intron fragment of Cox4i1. A luciferase vector containing p21RE was used as positive control. (G) State III oxygen consumption in HCT116 cells treated with siRNA control or two independent siRNAs targeting Cox4i1. Mean ± SD. (H) MitoSox Red staining of O2− in HCT116 treated as in G. (I) HCT116 cells treated as in G were challenged with H2O2, and cell death was measured by PI staining. Mean ± SD. (J) Wild-type MEFs treated with two different shRNAs against Cox4i1 (sh1 and sh2) or scramble control (scr) were assessed for expression of senescence markers at late passage (P7). Normalized expression levels are reported below the relative blots. (K) State III oxygen consumption in wild-type and knockout MEFs (P3) transfected for 36 h with empty vector or a plasmid expressing Flag-tagged mouse Cox4i1. State III was induced by injection of ADP. Expression of exogenous Cox4i1 was detected with anti-Flag antibody.
To ascertain whether TAp73-mediated regulation of Cox4i1 could be responsible for the mitochondrial dysfunction observed in TAp73-depleted cells, we knocked down Cox4i1 in HCT116 cells using two independent siRNAs. Depletion of Cox4i1 reduced oxygen flux (Fig. 5G) and increased anion superoxide levels (Fig. 5H). Recent findings suggest that mitochondrial respiration per se protects against oxygen-associated damage, and inhibition of mitochondrial activity elicits oxidative stress in cultured cells (Sung et al. 2010). In agreement with this hypothesis, Cox4 knockdown augmented H2O2-induced cell death (Fig. 5I), and, importantly, knockdown of Cox4i1 in primary wild-type MEFs also reproduced the senescent phenotype seen in the TAp73 knockout MEFs (Fig. 5J). Finally, ectopic expression of Cox4i1 in TAp73 knockout MEFs restored oxygen consumption to levels comparable with wild-type cells (Fig. 5K). Overall, these data suggest that Cox4i1 is a direct p73 target able to contribute to the mitochondrial impairment and oxidative imbalance triggered by depletion of TAp73.
The mechanisms that govern aging are still unclear and under intense investigation (Kenyon 2010). One widely accepted theory proposes that progressive accumulation of oxidative damage, arising from endogenously produced ROS, leads to tissue degeneration and age-related disorders (Balaban et al. 2005). However, the degree to which free radicals contribute to aging has recently been questioned (Lapointe and Hekimi 2010), although it is becoming increasingly evident that mitochondrial bioenergetics do play a crucial role in the aging process. Indeed, mitochondrial respiration decreases with aging (Yen et al. 1989). More directly, mice engineered to express an error-prone mutant mitochondrial polymerase δ show reduced longevity (Trifunovic et al. 2004), which correlates with decreased mitochondrial function but not with increased ROS production, suggesting that impaired metabolism resulting from compromised mitochondrial respiration may account for the aging phenotype (Trifunovic et al. 2005). Moreover, mitochondrial targeted overexpression of the scavenging enzyme catalase extends life span at least partially by preventing the age-associated decline in mitochondrial activity (Lee et al. 2010). While we cannot exclude the possibility that TAp73 influences aging by other mechanisms, compromised complex IV activity with increased ROS generation is clearly one mechanism by which TAp73 deficiency leads to an aging phenotype.
Dissociation between aging, insulin resistance, and ROS has been suggested from several models, and thus, under certain conditions, ROS may promote aging independently from its effects on glucose metabolism. Our finding that TAp73 knockout mice on a long-term HFD showed relatively increased insulin sensitivity could therefore be due to the positive effects of ROS on insulin signaling, as reported in JunD or GPX-1 knockout mice (Laurent et al. 2008; Loh et al. 2009).
The p53 family members p53 and p63 are pivotal in the control of organismal aging, although the underlying mechanisms are still unclear (Keyes et al. 2005; Matheu et al. 2008; Su et al. 2009; Sahin et al. 2011), and p53-dependent mitochondrial dysfunction has been described in aging-prone telomerase-deficient mice (Sahin et al. 2011) and age-related cardiovascular and adipose tissue changes (Minamino et al. 2009). Here we report that selective TAp73 depletion accelerates aging in vivo associated with altered mitochondrial function at least partially due to depletion of Cox4i1, and Cox4i1 knockdown itself results in accelerated senescence in vitro. Overall, our data are consistent with recent findings from in vivo mouse models of mitochondrial dysfunction and unbalanced ROS homeostasis (Trifunovic et al. 2004; Laurent et al. 2008; Chen et al. 2009; Liu et al. 2009; Loh et al. 2009; Reilly et al. 2010; Sahin et al. 2011) and expand the p53 and mitochondrial axis of aging with the addition of new players, TAp73 and Cox4i1.
Materials and methods
Mice
Generation and genotyping of TAp73 mice were described in Tomasini et al. (2008). Serum IGF was quantified by ELISA (R&D Systems). Body fat was measured by a PIXImus Small Animal Densitometer (Lunar). For the hair growth assay, age-matched wild-type and TAp73 knockout mice (18 mo) were shaved on their dorsal surface using an electric razor. Pictures were taken 24 d after shaving. Indirect calorimetry was performed using LabMaster (TSE Systems). Mice were acclimatized for 24 h before measurements were taken every 15 min for 24 h. Oxygen consumption (VO2) is expressed as milliliters of O2 consumed per kilogram of body weight per minute. Activity was expressed by movement-induced beam interruptions per minute in the calorimeter. Daily food intake was monitored by weighing food hoppers. For the diet-induced obesity model, mice were fed a HFD (60% of calories from fat; Research Diets) or a ND (standard chow, 10% calories from fat; GLP) for 16 wk, starting in 8-wk-old mice. Metabolic tests were previously described (Menghini et al. 2009).
Animals were treated in accordance with the NIH Guide for Care and Use of Laboratory Animals as approved by the Ontario Cancer Institute Animal Care Committee. Mice were bred and subjected to procedures under the project license released from the Home Office.
ROS measurement
To determine ROS levels, HCT116 cells and MEFs were stained for 10 min with 10 μM DCFDA (Invitrogen, M36008) in Hank's balanced salt solution (HBSS). To measure cellular anion superoxide content, HCT116 cells and MEFs were stained for 10 and 30 min, respectively with 1 μM MitoSox Red (Invitrogen, C6827) in HBSS. After washing with HBSS, samples were analyzed by flow cytometry, acquiring a minimum of 10,000 cells per sample. Each experiment was performed in triplicate and analyzed with CELLQuest software.
Respirometry and NADH
Oxygen consumption was measured at 37°C by high-resolution respirometry using Oxygraph-2k and DataLab software (Oroboros). Cells and tissues were homogenized in respiration medium (0.5 mM EGTA, 3 mM MgCl2·6 H2O, 60 mM K-lactobionate, 20 mM taurine, 10 mM KH2P04, 20 mM HEPES, 110 mM sucrose, 1g/L BSA) and assayed with 2-sec-interval measurements. State III respiration was induced by addition of 2 mM malate, 10 mM glutamate, and 5–10 mM ADP.
NADH was measured spectroscopically in protein extracts (excitation wavelength 366 nm; absorbance wavelength 460 nm) and normalized to protein content.
Acknowledgments
We thank David J. Read, Jenny Edwards, and Lucia Pinon (the Medical Research Council [MRC], UK, Toxicology Unit) . We thank Alessandro Terrinoni (the Immaculate Dermopatico Institute, Istituto Dermopatico dell'Immacolata, Istituto di Ricovero e Cura a Carattere Scientifico [IDI-IRCCS]) for help with the ChIP protocol, and the Samuel Lunenfeld Research Institute's Centre for Modeling Human Disease Mouse Phenotyping Facility for their screening services (http://www.cmhd.ca). This work was supported by the MRC, UK; the Associazione Italiana per la Ricerca sul Cancro (AIRC) [(2008-2010_33-08) (#5471) (2011-IG11955)]; AIRC 5xmille (#9979); and Telethon Grant GGPO9133 to G.M. Research described here was also supported in part by the Ministero della Salute (Ricerca oncologica 26/07) and IDI-IRCCS (RF06 c.73, RF07 c.57, RF08 c.15, and RF07 c.57) to G.M.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.197640.112.
References
- Balaban RS, Nemoto S, Finkel T 2005. Mitochondria, oxidants, and aging. Cell 120: 483–495 [DOI] [PubMed] [Google Scholar]
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ 2010. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol 2: a001198 doi: 10.1101/cshperspect.a001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Kao CH, Chen YT, Wang CH, Wu CY, Tsai CY, Liu FC, Yang CW, Wei YH, Hsu MT, et al. 2009. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev 23: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Vousden KH 2010. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol 2: a001040 doi: 10.1101/cshperspect.a001040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ 2010. The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA 2005. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev 19: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE 2004. Ink4a/Arf expression is a biomarker of aging. J Clin Invest 114: 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S 2010. When a theory of aging ages badly. Cell Mol Life Sci 67: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G, Solari F, Mateescu B, Karaca M, Castel J, Bourachot B, Magnan C, Billaud M, Mechta-Grigoriou F 2008. Oxidative stress contributes to aging by enhancing pancreatic angiogenesis and insulin signaling. Cell Metab 7: 113–124 [DOI] [PubMed] [Google Scholar]
- Lee HY, Choi CS, Birkenfeld AL, Alves TC, Jornayvaz FR, Jurczak MJ, Zhang D, Woo DK, Shadel GS, Ladiges W, et al. 2010. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab 12: 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G 2011. The p53 family: Guardians of maternal reproduction. Nat Rev Mol Cell Biol 12: 259–265 [DOI] [PubMed] [Google Scholar]
- Lin YL, Sengupta S, Gurdziel K, Bell GW, Jacks T, Flores ER 2009. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet 5: e1000680 doi: 10.1371/journal.pgen.1000680 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. 2009. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K, Deng H, Fukushima A, Cai X, Boivin B, Galic S, Bruce C, Shields BJ, Skiba B, Ooms LM, et al. 2009. Reactive oxygen species enhance insulin sensitivity. Cell Metab 10: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Serrano M 2008. The Arf/p53 pathway in cancer and aging. Cancer Res 68: 6031–6034 [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM 2006. p53 regulates mitochondrial respiration. Science 312: 1650–1653 [DOI] [PubMed] [Google Scholar]
- Melino G, De Laurenzi V, Vousden KH 2002. p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2: 605–615 [DOI] [PubMed] [Google Scholar]
- Menghini R, Menini S, Amoruso R, Fiorentino L, Casagrande V, Marzano V, Tornei F, Bertucci P, Iacobini C, Serino M et al. 2009. Tissue inhibitor of metalloproteinase 3 deficiency causes hepatic steatosis and adipose tissue inflammation in mice. Gastroenterology 136: 663–672.e4 doi: 10.1053/j.gastro.2008.10.079 [DOI] [PubMed] [Google Scholar]
- Minamino T, Orimo M, Shimizu I, Kunieda T, Yokoyama M, Ito T, Nojima A, Nabetani A, Oike Y, Matsubara H, et al. 2009. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med 15: 1082–1087 [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SM, Bhargava P, Liu S, Gangl MR, Gorgun C, Nofsinger RR, Evans RM, Qi L, Hu FB, Lee CH 2010. Nuclear receptor corepressor SMRT regulates mitochondrial oxidative metabolism and mediates aging-related metabolic deterioration. Cell Metab 12: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA 2012. Axis of ageing: Telomeres, p53 and mitochondria. Nat Rev Mol Cell Biol 13: 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. 2011. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, et al. 2009. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HJ, Ma W, Wang PY, Hynes J, O'Riordan TC, Combs CA, McCoy JP Jr, Bunz F, Kang JG, Hwang PM 2010. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun 1: 5 doi: 10.1038/ncomms1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. 2008. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 22: 2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423 [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG 2005. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci 102: 17993–17998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, et al. 2010. Isoform-specific p73 knockout mice reveal a novel role for ΔNp73 in the DNA damage response pathway. Genes Dev 24: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404: 99–103 [DOI] [PubMed] [Google Scholar]
- Yen TC, Chen YS, King KL, Yeh SH, Wei YH 1989. Liver mitochondrial respiratory functions decline with age. Biochem Biophys Res Commun 165: 944–1003 [DOI] [PubMed] [Google Scholar]