Different cyclin types have distinct abilities to reverse the S-phase checkpoint, and timely entry into mitosis after embryonic S phase requires collaborative action of multiple cyclin types.
Abstract
Precise timing coordinates cell proliferation with embryonic morphogenesis. As Drosophila melanogaster embryos approach cell cycle 14 and the midblastula transition, rapid embryonic cell cycles slow because S phase lengthens, which delays mitosis via the S-phase checkpoint. We probed the contributions of each of the three mitotic cyclins to this timing of interphase duration. Each pairwise RNA interference knockdown of two cyclins lengthened interphase 13 by introducing a G2 phase of a distinct duration. In contrast, pairwise cyclin knockdowns failed to introduce a G2 in embryos that lacked an S-phase checkpoint. Thus, the single remaining cyclin is sufficient to induce early mitotic entry, but reversal of the S-phase checkpoint is compromised by pairwise cyclin knockdown. Manipulating cyclin levels revealed that the diversity of cyclin types rather than cyclin level influenced checkpoint reversal. We conclude that different cyclin types have distinct abilities to reverse the checkpoint but that they collaborate to do so rapidly.
Introduction
Most metazoan species lay large eggs with provisions for the entirety of embryogenesis. These eggs begin embryogenesis with rapid cell cycles, and when cell number is adequate to begin development, the cell cycles slow, and zygotic gene expression from the newly amplified nuclei begins to direct the events of morphogenesis (O’Farrell et al., 2004). This transition from rampant proliferation to morphogenesis is called the midblastula transition (MBT; Newport and Kirschner, 1982a,b). Throughout these stages, the mechanisms timing the cell cycle and those timing development are interwoven. We are probing the basis of this temporal control.
Drosophila melanogaster embryogenesis begins with 13 rapid, synchronous mitotic cycles that occur without zygotic gene expression. These cycles lack gap phases and cytokinesis. Instead, they oscillate between S phase and mitosis, amplifying nuclei in a syncytial cytoplasm until the MBT, which occurs in interphase of cycle 14. Early interphases extend progressively, beginning as short as 3.4 min, lengthening to 12 min by cycle 13, and abruptly jumping to 90 min or more in cycle 14, the first asynchronous cycle (Foe and Alberts, 1983; Edgar et al., 1986; Shermoen et al., 2010). This temporal course is tightly coupled with development so that cycle 14 is marked by cellularization of the syncytial nuclei and onset of gastrulation.
Usually, mitosis and progress to the next cycle are triggered by Cdc25 phosphatase’s removal of inhibitory phosphate from preformed cyclin–Cdk1 complexes (Russell and Nurse, 1986; Edgar and O’Farrell, 1989; O’Farrell, 2001). Indeed, at the first post-MBT mitosis, cyclins are in excess in Drosophila, and a pulse of transcription of the string gene, which encodes a Cdc25 phosphatase, times mitosis (Edgar and O’Farrell, 1989, 1990; Lehner and O’Farrell, 1989; Edgar et al., 1994). However, the pre-MBT cycles are independent of transcription, and so, their timing cannot be governed by Cdc25 transcription.
Although it has been suggested that accumulation of cyclin times early rapid embryonic cycles (Murray and Kirschner, 1989), studies in Drosophila have instead implicated S-phase duration as the interphase timer. In the earliest S phases, the genome is replicated remarkably quickly by the simultaneous firing of many origins. During cycles 11 to 13, slight but increasing delays in the onset of replication of heterochromatic satellite sequences gradually extend S phase (McCleland et al., 2009a; Shermoen et al., 2010). During these cycles, complete deletion of S phase, by blocking the formation of prereplication complexes, shortens interphase, indicating that S phase indirectly or directly times interphase duration (McCleland et al., 2009a). Mutations that inactivate the S-phase checkpoint, mei-41 (dATR) or grapes (grp; dChk1), also shorten interphase and, by the time of the thirteenth cycle, result in catastrophe when nuclei enter mitosis with incompletely replicated DNA (Sibon et al., 1997, 1999; Yu et al., 2000). Thus, gradually lengthening S phase acts through the S-phase checkpoint to govern interphase length.
Some experiments that altered maternal cyclin gene dose showed a cyclin influence on the length of early cycles (Edgar et al., 1994; Stiffler et al., 1999). These findings were initially interpreted as an indication that cyclin levels time mitotic entry; however, we would now like to test alternative interpretations that might be more consistent with findings implicating S phase as the interphase timer. Like most organisms, Drosophila has multiple mitotic cyclins—CycA, CycB, and CycB3—which exhibit partial redundancy (Jacobs et al., 1998). Consistent with redundancy, RNAi knockdown of all three cyclins was required to arrest early embryos in interphase (McCleland and O’Farrell, 2008). However, unique defects that were seen when a single cyclin promoted mitosis revealed that the different cyclin types differ in their action (McCleland et al., 2009b). Because a change in gene dose of a cyclin will alter the relative abundance of cyclin types as well as cyclin level, the consequences might be caused by either of these changes. We have made an effort to distinguish cyclin level and cyclin-type effects on the cell cycle.
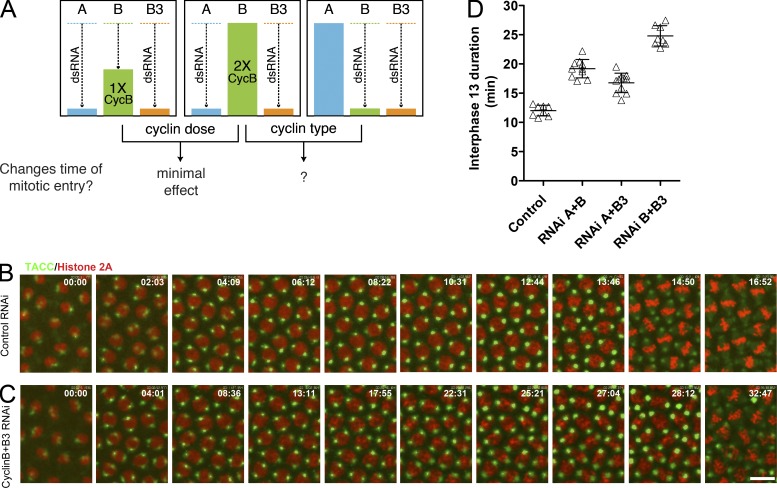
Previously, we tested the influence of cyclin level without the obfuscation of changing distributions of cyclin type; two of the three mitotic cyclins were knocked down by RNAi, and the level of expression of the one remaining cyclin was altered by changing gene dose. In this situation, change in the gene dose of the remaining cyclin did not substantially alter interphase duration (Fig. 1 A), indicating that the accumulation of the cyclin protein does not limit entry into mitosis in this experimental situation (McCleland et al., 2009b). This finding suggested that cyclin levels are in excess of requirements for mitotic entry, a conclusion in accord with the finding that S-phase duration governs interphase length (Sibon et al., 1997, 1999; McCleland et al., 2009a).
Figure 1.
Cyclin-type effect on interphase timing. (A) As illustrated for RNAi knockdown of CycA (A) and CycB3 (B3), after pairwise cyclin knockdown, the dose of the remaining cyclin (CycB, B) did not significantly affect interphase length (McCleland et al., 2009b). Halving the gene dose of the remaining cyclin (performed for all combinations) had a minimal effect on interphase length (comparison on the left), arguing that cyclin level is not a major determinant of interphase length in these conditions. In contrast, the type of cyclin remaining (comparison on the right) influences interphase duration (this study). (B and C) Real-time records of histone (RFP-H2AvD) and centrosomes (GFP-TACC) show that RNAi against CycB+B3 (C), which created a situation in which the cell cycle was running mainly on CycA, greatly extended the interphase length in cycle 13 compared with the control (B; 28:12 vs. 13:46 [minutes and seconds]). Bar, 10 µm. (D) Interphase durations in control and each of the pairwise cyclin knockdown embryos. Mean interphase 13 length was 12.02 ± 0.92 min in control embryos, whereas knockdown of CycA+B, CycA+B3, or CycB+B3 extended interphase to 19.19 ± 1.57, 16.76 ± 1.67, or 24.81 ± 1.75 min, respectively (±SD). Horizontal lines show means.
Thus, the past observations might be explained if changes in the distributions of cyclin type influenced interphase duration. This could be understood if the cyclins differed in their potency to trigger mitosis. For example, it was proposed that CycA could be specialized to prime entry into mitosis (Clarke et al., 1992; Gong et al., 2007). Alternatively, because we recently showed that down-regulation of all three “mitotic” cyclins in the early cycles extends S phase (Farrell et al., 2012), perhaps the cyclin types could differ in abilities to accelerate S phase. Surprisingly, however, the cyclin-type input does not involve either of these mechanisms.
We knocked down two cyclins and then evaluated the ability of the remaining one to support interphase 13 progression. Each pairwise knockdown prolonged interphase to a different degree (cyclin-type effect). In the prolonged interphase, S phase was barely extended, and instead, a gap phase was introduced after S phase. Different gap-phase durations accounted for the cyclin-type effect. Inactivation of the DNA replication checkpoint eliminated this new gap phase, restored a short interphase, and removed the cyclin-type effect on interphase duration. Thus, the G2 introduced by cyclin knockdown requires the S-phase checkpoint, indicating that the G2 represents the time of recovery from the checkpoint. Consequently, the cyclin-type effect reflects differences in the ability of the different cyclins to promote checkpoint reversal.
Results and discussion
Cyclin type influences interphase length more than cyclin level
RNAi of all three mitotic cyclins in the syncytial embryo arrests the cycle in an interphase that exhibits a prolonged S phase and, before cycle 13, uncoupled centrosome replication (McCleland and O’Farrell, 2008; Farrell et al., 2012). In contrast, pairwise knockdown of cyclins allows progress to mitoses that exhibit distinctive defects depending on which cyclin remains (McCleland et al., 2009b). Here, we examine the consequence of pairwise knockdown of cyclins on interphase time.
Two of the three cyclins were removed by double-strand RNA (dsRNA) injection, and the cycle driven by the remaining cyclin was evaluated by live imaging. For example, CycB and CycB3 (abbreviated as CycB+B3 hereafter) were knocked down, and progress through the cycle with the remaining CycA (Fig. 1 C) was compared with control (Fig. 1 B). Transforming acidic coiled coil (TACC)–GFP and H2AvD-RFP were used to visualize the centrosome and nuclear cycles, respectively. Interphase duration was extended after knockdown of any pair of cyclins (Fig. 1, C and D). The centrosome cycle was extended in coordination with the cell cycle (Fig. 1 C). The results show that any single remaining cyclin is sufficient to promote mitosis 13 and a coordinated centrosome cycle but that no single cyclin can support normal interphase timing.
The prolongation of interphase upon pairwise knockdown suggested that cyclins have a role in defining interphase length. In previous work, we halved the level of the remaining cyclin by reducing gene dose in embryos with two knocked down cyclins (Fig. 1 A). This reduction of cyclin level had little effect on interphase timing, but it compromised execution of mitotic events (McCleland et al., 2009b). Thus, after pairwise cyclin knockdown, cyclin levels limit execution of mitosis but not the timing of mitotic entry.
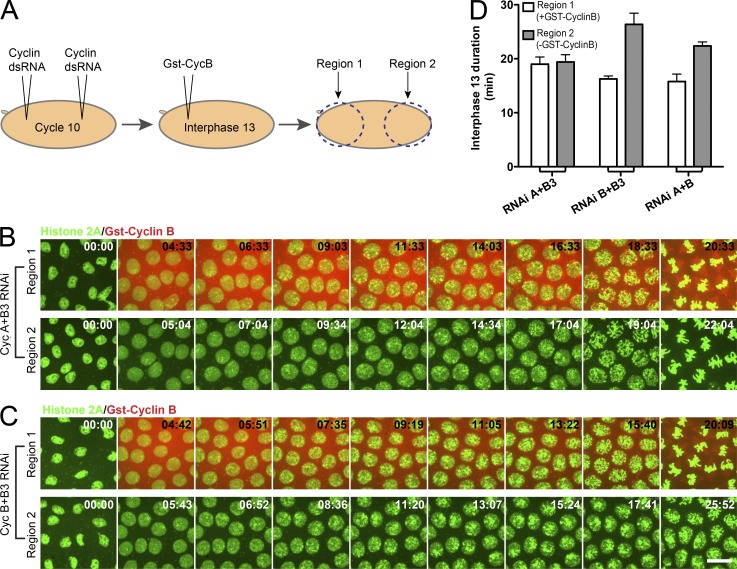
In addition to the published reduction of function experiments, we wanted to test whether increasing the level of cyclin would reveal a cyclin level input into the timing of mitosis. To this end, we injected embryos with different pairwise combinations of cyclin dsRNA, and when they reached cycle 13, we injected them with a purified Drosophila GST-CycB fusion protein at one pole (Fig. 2 A). In embryos running on CycA alone (CycB+B3 RNAi) or embryos running on CycB3 alone (CycA+B RNAi), the injected GST-CycB accelerated progress to mitosis (Fig. 2, C and D; and Videos 2 and 3). However, in embryos running on CycB alone (CycA+B3 RNAi), the injected GST-CycB did not accelerate progress to mitosis (Fig. 2, B and D; and Video 1). Thus, injected CycB synergized with CycA or CycB3 to advance mitosis, but the same injection of CycB did not advance mitosis in embryos in which endogenous CycB was the remaining cyclin. Therefore, restoration of a second cyclin type, but not an increase in cyclin level, advanced mitosis. This experiment supports previous gene dose experiments in arguing that the level of the single remaining cyclin is not a major determinant of interphase length under the pairwise cyclin knockdown conditions. We propose that the different cyclin types have somewhat specialized activities and collaborate to promote rapid entry into mitosis. We have sought an explanation for this phenomenon and have uncovered an unexpected influence of cyclin type on cell cycle progress.
Figure 2.
Diversity of cyclin types influences interphase length, whereas amount of a single cyclin has little effect. (A) Schematic of the experiment. dsRNA to two of the three cyclins was introduced throughout the embryo in cycle 10. Rhodamine-tagged GST-CycB protein was then injected at one pole during interphase 13. Embryos were imaged in regions 1 and 2, and the timing of mitosis in the two regions was determined. (B and C) Video frames of cycle 13 in which the remaining cyclin is CycB (B) or CycA (C). After injection of the GST-CycB protein, red fluorescence is seen in the injected pole (rhodamine). The absence of red at the other pole shows that significant GST-CycB does not reach the other pole in this time frame. Noting time stamps (minutes and seconds) on the images, it can be seen that GST-CycB does not advance mitosis when introduced into the CycB alone embryo but does advance mitosis when introduced into the CycA alone embryo. Bar, 5 µm. (D) A compilation of timing results from these experiments. Error bars represent SDs. n = 3.
Pairwise cyclin knockdown introduces a gap phase
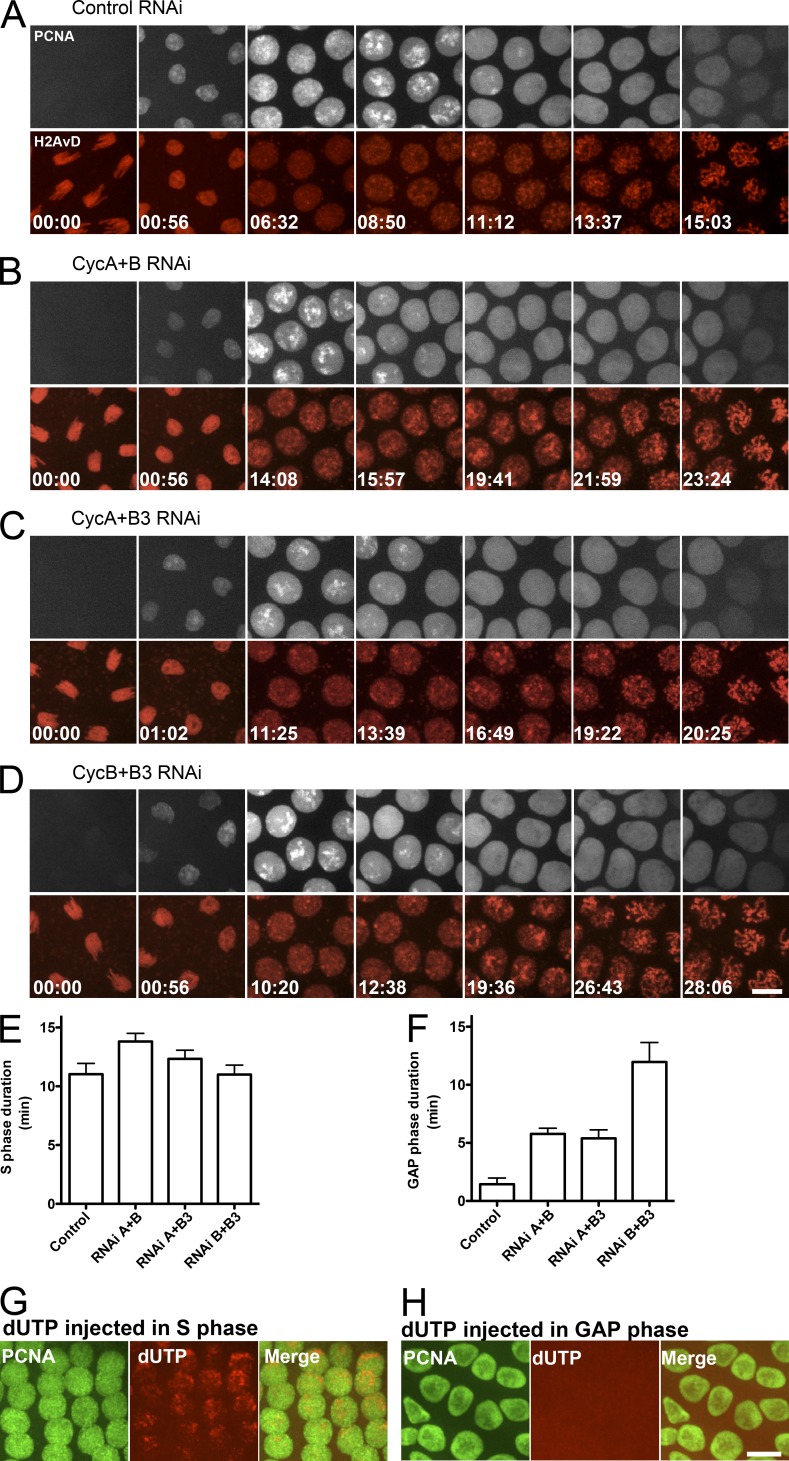
Because S-phase duration governs interphase length in the early cycles (McCleland et al., 2009a) and we recently showed that cyclin–Cdk1 activity shortens S phase (Farrell et al., 2012), we tested whether pairwise cyclin knockdown extends S phase. The changing distributions of proliferating cell nuclear antigen (PCNA)–GFP, which marks the sliding clamp of DNA polymerase, allowed us to visualize S phase (McCleland et al., 2009a; Farrell et al., 2012). We injected pairwise combinations of cyclin dsRNA into embryos expressing H2AvD-RFP and then injected PCNA-GFP recombinant protein and imaged cells near the point of injection. In control embryos (Fig. 3 A and Video 4), PCNA accumulated in the nucleus at the onset of interphase (Fig. 3 A, 00:56) and then became increasingly restricted to foci that dimmed and dispersed at the end of S phase (Fig. 3 A, 11:12). Dispersal of PCNA from the nucleus marked nuclear envelope breakdown (Fig. 3 A, 13:37 and 15:03). Except for a delay in nuclear envelope breakdown, the PCNA dynamics showed little change upon pairwise cyclin knockdown (Fig. 3, B–D; and Videos 5, 6, and 7), and the extension of S phase was small in comparison to the change in interphase (Fig. 3 E). Additionally, the S phase extension did not approach the near doubling of S phase caused by the triple cyclin knockdown (Farrell et al., 2012).
Figure 3.
Interphase progression in cyclin RNAi-treated embryos. (A–D) Video frames of GFP-PCNA (top; white) and histone-RFP (bottom; red) during the thirteenth syncytial cycle. Time is given in minutes and seconds. (A) Control embryos had a short gap phase before mitotic entry (<2 min, between 11:12 and 13:37; Video 1). (B–D) Pairwise knockdown of mitotic cyclins prolonged interphase, mainly by extending the G2 (e.g., ∼6 min between 15:57 and 21:59 in B; Video 2). (E) S-phase duration. Mild prolongations of S phase were observed in CycA+B and CycA+B3 RNAi-treated embryos as compared with the control, whereas treatment with CycB+B3 RNAi had no significant effect. (F) Gap-phase duration. All the combinations of cyclin RNAi treatment extended the gap phase. CycB+B3 RNAi was most dramatic. Error bars represent SDs. n > 3. (G and H) Alexa Fluor 546–dUTP (red) was incorporated into DNA when injected before but not after the dispersal of PCNA foci (green) in CycB+B3 RNAi-treated embryos. Bars, 5 µm.
Pairwise cyclin knockdown introduced a distinct pause between the completion of S phase and onset of chromosome condensation (Fig. 3, B–D and F). To confirm that PCNA foci correctly marked S phase and that DNA replication did not extend into the “gap” phase, Alexa Fluor 546–labeled deoxy-UTP (dUTP) was injected into these cyclin RNAi-treated embryos before and after the dispersal of PCNA foci. For all knockdowns, nucleotide was incorporated before, but not after, PCNA foci dispersal (e.g., Fig. 3, G and H), indicating that PCNA appropriately marks completion of active replication.
The finding that no pairwise cyclin knockdown gave the dramatic extension of S phase previously reported after triple cyclin knockdown (Farrell et al., 2012) leads us to conclude that each cyclin type is capable of accelerating S phase. Furthermore, we conclude that pairwise cyclin knockdown in the syncytial cycles generates a G2-like gap phase whose duration depends on which cyclins are knocked down (Fig. 3 F).
The induced G2 depends on the DNA replication checkpoint
Because cyclin levels are not limiting, we presumed that something else must be responsible for the G2 that is created upon knockdown of two of the three cyclins. How can this be?
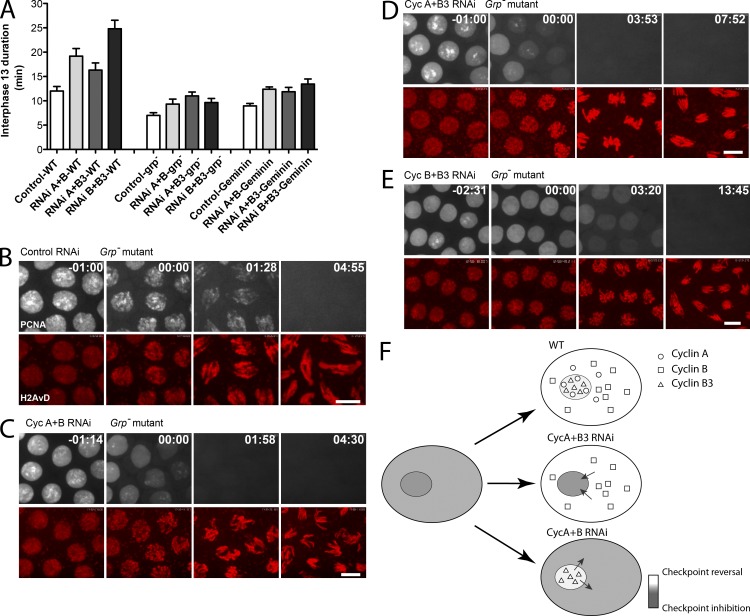
We tested the contribution of S phase by deleting S phase to examine the consequence on the time of mitotic entry. Injection of the Cdt1 inhibitor Geminin blocks the licensing of replication origins, thereby deleting the subsequent S phase (McCleland et al., 2009a). As previously reported, Geminin injection into cycle 12 control embryos, which deletes S phase 13, shortened interphase 13. Geminin injection also shortened interphase 13 in the cyclin knockdown embryos, shortening it almost to the duration of interphase in embryos with a full complement of cyclins (Fig. 4 A). S phase deletion also attenuated the differences in interphase length among the different cyclin knockdowns. Thus, S phase influences interphase duration, even though it is ordinarily completed well before mitosis in the cyclin knockdown embryos. Furthermore, the different cyclin types drive mitosis at similar times in the absence of S phase, suggesting similar potencies to drive mitosis under these conditions.
Figure 4.
The G2 and prolonged interphase after pairwise cyclin knockdown requires the replication checkpoint. (A) Inactivation of Chk1 (grp−) or deletion of S phase (by Geminin injection) shortened interphase in control and cyclin RNAi-treated embryos and reduced the differences among cyclin types. Error bars represent SDs. Detailed measurements are listed in Table S1. Data for cyclin RNAi experiments in wild-type embryos are reproduced from Fig. 1 D for purpose of comparison. (B–E) Video frames of grp− mutant embryos in cycle 13 (GFP-PCNA is shown in white, and histone-RFP is shown in red) aligned at the start of DNA condensation (t = 00:00). Time is given in minutes and seconds. Bars, 5 µm. (F) A schematic model in which checkpoint inhibition of cyclin–Cdk1 (gray) is reversed by compartment-specific action of cyclins plus a slow communication between compartments (arrows).
Pairwise knockdown of cyclins in embryos lacking Chk1/Grapes (embryos from grp mutant mothers) modestly extends interphase in the grp embryo (Fig. 4 A). This knockdown also substantially suppresses the mitosis 13 defects in grp embryos (Figs. 4, B–E; and S2, A and B). The mitotic catastrophe in grp embryos has long been thought to be caused by entry into mitosis with incompletely replicated DNA. Indeed, our analysis of PCNA localization (Figs. 4 and S2, C and D) supports a proposal that the small extension of interphase allows completion of S phase and, hence, suppression of the catastrophe. This suppression of the grp phenotype, which extends to partial restoration of gastrulation (Video 8), is reminiscent of suppression of hypomorphic mei-41 mutations when the maternal dose of CycA and/or CycB was reduced (Sibon et al., 1999). However, the most important feature of this analysis is that the extension of interphase by pairwise cyclin knockdown in grp embryos is so slight that interphase remains shorter than a wild-type interphase 13 (Fig. 4 A). Thus, each individual cyclin type can drive rapid advance to mitosis in the absence of functional Chk1. Furthermore, the G2 that was introduced by cyclin knockdown was absent in grp embryos (Fig. 4, C–E), and cyclin type–specific differences in interphase length were minor (Fig. 4 A). These results demonstrate that mitotic entry is timed primarily by the Grapes-dependent checkpoint in cyclin knockdown embryos and, moreover, that the G2 induced in these embryos results from action of the checkpoint.
How might S phase govern the time of mitosis when mitosis begins well after completion of S phase? To test whether the S-phase checkpoint delayed accumulation of the remaining cyclin, we immunoblotted single knockdown embryos to follow accumulation in wild-type and grp embryos. Cyclin accumulated during S phase in both wild-type and grp mutant embryos (Fig. S3). Thus, Grapes function does not delay the production of cyclin. Instead, the difference between grp+ and grp− embryos suggests that persistent activity of the checkpoint prevents the checkpoint-competent embryos from going into mitosis after pairwise cyclin knockdown. Because wild-type embryos enter mitosis immediately after S phase, the checkpoint is rapidly reversed when there is a full complement of cyclin types, but its reversal is delayed upon pairwise cyclin knockdown. Apparently, the different cyclin types ordinarily collaborate to rapidly reverse the checkpoint.
Compartments and cyclin specialization
Our findings show that a persistent action or consequence of the DNA replication checkpoint underlies a G2 phase that is introduced by pairwise knockdown of cyclins. One might propose that cyclin knockdown doesn’t really cause persistence of the checkpoint activity but simply makes a slowly decaying checkpoint function longer by compromising the cyclin–Cdk1 activity that must be suppressed by the checkpoint. We disfavor such an interpretation because it is quantitative, and the data argue that neither reduction nor increase in the remaining cyclin affects the duration of interphase. Instead, we argue that cyclin–Cdk1 contributes to shutting off the checkpoint and propose that efficient shutoff of the checkpoint requires multiple cyclin types. One way to explain this is based on the distinct subcellular localizations of mitotic cyclins (Jacobs et al., 1998; Stiffler et al., 1999). Once activated, the checkpoint can operate in multiple cellular compartments, such as the nucleus and the cytoplasm. Although signals coordinate entry into mitosis in the cytoplasm and nucleus (Gavet and Pines, 2010), persistent nuclear checkpoint activity can prevent mitotic entry despite cytoplasmic Cdk activity (Heald et al., 1993). Individual cyclins would not be able to act on their own to reverse the checkpoint in all compartments if each is excluded from one compartment. For example, cyclin B is efficiently excluded from the nucleus in cycle 13 embryos (Maldonado-Codina and Glover, 1992) and presumably would not contribute to checkpoint reversal in this compartment, whereas cyclin B3 is nuclear (Jacobs et al., 1998). In embryos with only CycB, the checkpoint should be reversed first in the cytoplasm; however, progress to mitosis should depend on slower reversal in the nucleus, which might be based on communication between compartments (Fig. 4 F). Consistent with this proposal, injection of CycB protein preferentially drove cytoplasmic, but not nuclear, mitotic events (Royou et al., 2008). The full complement of cyclins with distinct localizations, however, appears to reverse the checkpoint promptly and coordinately in all the compartments.
Our data demonstrate a cyclin-type effect on reversal of the DNA replication checkpoint, which emphasizes the qualitatively distinct contributions among mitotic cyclins during mitotic entry. This study opens many further questions, such as what causes the checkpoint to inactivate? How do mitotic cyclins promote checkpoint reversal? We believe answers to these questions will help us fully understand the timing mechanism of the cell division cycle.
Materials and methods
Fly stocks
Strains of Drosophila were maintained on standard cornmeal–yeast medium. The following stocks were used for live-embryo imaging: (a) w; ; H2AvD-GFP/TM6, Tb, (b) w; ; H2AvD-RFP/TM6, Tb, (c) w; UASp-YFP-PCNA/CyO; Gal4-VP16/MKRS, and (d) w, TACC-GFP; ; H2AvD-RFP/TM6, Tb (Clarkson and Saint, 1999; McCleland and O’Farrell, 2008; McCleland et al., 2009a,b). Stock w; grp06034, H2AvD-GFP was used in the DNA replication checkpoint–deficient experiments (McCleland et al., 2009a).
Microinjection and live-embryo imaging
Cyclin dsRNA was made with T7 RNA polymerase, as detailed in McCleland and O’Farrell (2008). GST- or His-tagged recombinant proteins used in the experiments were made previously (Royou et al., 2008; McCleland et al., 2009a; Shermoen et al., 2010). Briefly, they were produced in BL-21 DE3 pLysS bacteria (Agilent Technologies) and purified on glutathione–Sepharose (GE Healthcare) or nickel agarose beads (QIAGEN) according to the manufacturer’s instructions.
Embryos were collected and treated as previously described (Farrell et al., 2012). Briefly, embryos were collected on grape–agar plates for 30 min and then aged for 25 min at 25°C. They were then dechorionated for 2 min in 50% bleach, aligned, and taped onto a coverslip, desiccated for 8–9 min, and covered in halocarbon oil (Sigma-Aldrich) for injection.
For the dsRNA injections, embryos were aged to cycle 9 before injection to get a robust and consistent knockdown efficiency. Cyclin dsRNAs were dissolved in a buffer containing 0.25 mM potassium phosphate, pH 7.4, and 2.5 mM KCl and injected at the concentration of 0.5 mg/ml. For GST-CycB injections, rhodamine-labeled recombinant protein (a gift from W. Sullivan, University of California, Santa Cruz, Santa Cruz, CA) was injected in early interphase 13 at the concentration of 31 µM. For Geminin protein injections, purified His-tagged Geminin (made by M. McCleland, Genentech, South San Francisco, CA) was dialyzed into 40 mM Hepes, pH 7.4, and 150 mM KCl and injected at 10 mg/ml in interphase 12 to delete the S phase in interphase 13. To visualize S phase, recombinant PCNA-GFP protein (made by M. McCleland) was injected at the concentration of 2 mg/ml. Hydroxyurea was used at 200 mM.
For live-embryo experiments, embryos were covered with halocarbon oil and imaged with a spinning-disk confocal system (CSU10; Yokogawa Corporation of America) equipped with a high-resolution digital charge-coupled device camera (ORCA-ER; Hamamatsu Photonics), on an inverted microscope (DM IRB; Leica) with 100× Plan Fluotar 1.3 NA and 40× Plan Fluotar 0.7 NA objectives at room temperature. Most image stacks were collected at 1 µm over a 5-µm range using a controlled stage (MS-2000; Applied Scientific Instrumentation) at a time interval of 30 s. All images were captured and processed in Volocity 6 (PerkinElmer). Statistics were performed in Prism (GraphPad Software).
dUTP incorporation
Embryos expressing YFP-PCNA were first injected with cyclin dsRNA around cycle 9 and aged into interphase 13. Then, on-scope injection of 50 µM Alexa Fluor 546–dUTP (obtained from Invitrogen) was performed before or after disappearance of PCNA foci, which indicated S phase and the gap phase, respectively. After 3 min of incorporation, embryos were fixed in 37% formaldehyde (Thermo Fisher Scientific) for 10 min, hand devitellinized, and stained with GFP antibody (Invitrogen) and DAPI (Invitrogen).
Single-embryo Western blot
Wild-type or grp mutant embryos were injected with cyclin dsRNAs and then followed by live-embryo imaging. Embryos were then gently detached from the coverglass with an art brush at the indicated time in interphase 13, fixed in 1:1 methanol/heptane mixture on ice, vortexed with glass beads in 2× SDS sample buffer, and then boiled for 8 min. Rabbit anti-CycA antibody (made by C. Lehner [University of Zurich, Zurich, Switzerland] as detailed in Lehner and O’Farrell, 1989) was used at 1:1,000. Mouse anti-CycB antibody (F2F4; Developmental Studies Hybridoma Bank) was used at 1:5. Mouse anti-PSTAIRE (Sigma-Aldrich) was used at 1:4,000.
Online supplemental material
Fig. S1 evaluates the amount of injected GST-CycB relative to the level of endogenous CycB. Fig. S2 illustrates suppression of grp mutant phenotypes by cyclin knockdowns. Fig. S3 shows comparable cyclin accumulation in wild-type and grp mutant embryos. Table S1 lists additional information for Fig. 4 A. Video 1 shows that injection of GST-CycB does not accelerate mitotic entry in CycA+B3 RNAi-treated embryos. Videos 2 and 3 are time-lapse videos of interphase 13 after GST-CycB injection in different cyclin RNAi-treated embryos targeting CycB and CycB3 and CycA and CycB, respectively. Videos 4–7 show time-lapse videos of embryos in Fig. 3 (A–D) progressing through interphase 13. Video 8 shows gastrulation of a grp− mutant embryo after CycB+B3 knockdown. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201205007/DC1.
Supplementary Material
Acknowledgments
We thank members of the O’Farrell laboratory for technical assistance and helpful comments and William Sullivan, Mark McCleland, and Christian Lehner for reagents.
This research was supported by National Institutes of Health grant GM037193 to P.H. O’Farrell.
Footnotes
Abbreviations used in this paper:
- dsRNA
- double-strand RNA
- dUTP
- deoxy-UTP
- MBT
- midblastula transition
- PCNA
- proliferating cell nuclear antigen
- TACC
- transforming acidic coiled coil
References
- Clarke P.R., Leiss D., Pagano M., Karsenti E. 1992. Cyclin A- and cyclin B-dependent protein kinases are regulated by different mechanisms in Xenopus egg extracts. EMBO J. 11:1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson M., Saint R. 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18:457–462 10.1089/104454999315178 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., O’Farrell P.H. 1989. Genetic control of cell division patterns in the Drosophila embryo. Cell. 57:177–187 10.1016/0092-8674(89)90183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., O’Farrell P.H. 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 62:469–480 10.1016/0092-8674(90)90012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Kiehle C.P., Schubiger G. 1986. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 44:365–372 10.1016/0092-8674(86)90771-3 [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Sprenger F., Duronio R.J., Leopold P., O’Farrell P.H. 1994. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 8:440–452 10.1101/gad.8.4.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J.A., Shermoen A.W., Yuan K., O’Farrell P.H. 2012. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev. 26:714–725 10.1101/gad.186429.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V.E., Alberts B.M. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61:31–70 [DOI] [PubMed] [Google Scholar]
- Gavet O., Pines J. 2010. Activation of cyclin B1–Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189:247–259 10.1083/jcb.200909144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D., Pomerening J.R., Myers J.W., Gustavsson C., Jones J.T., Hahn A.T., Meyer T., Ferrell J.E., Jr 2007. Cyclin A2 regulates nuclear-envelope breakdown and the nuclear accumulation of cyclin B1. Curr. Biol. 17:85–91 10.1016/j.cub.2006.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., McLoughlin M., McKeon F. 1993. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 74:463–474 10.1016/0092-8674(93)80048-J [DOI] [PubMed] [Google Scholar]
- Jacobs H.W., Knoblich J.A., Lehner C.F. 1998. Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev. 12:3741–3751 10.1101/gad.12.23.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner C.F., O’Farrell P.H. 1989. Expression and function of Drosophila cyclin A during embryonic cell cycle progression. Cell. 56:957–968 10.1016/0092-8674(89)90629-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Codina G., Glover D.M. 1992. Cyclins A and B associate with chromatin and the polar regions of spindles, respectively, and do not undergo complete degradation at anaphase in syncytial Drosophila embryos. J. Cell Biol. 116:967–976 10.1083/jcb.116.4.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., O’Farrell P.H. 2008. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle. Curr. Biol. 18:245–254 10.1016/j.cub.2008.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., Shermoen A.W., O’Farrell P.H. 2009a. DNA replication times the cell cycle and contributes to the mid-blastula transition in Drosophila embryos. J. Cell Biol. 187:7–14 10.1083/jcb.200906191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., Farrell J.A., O’Farrell P.H. 2009b. Influence of cyclin type and dose on mitotic entry and progression in the early Drosophila embryo. J. Cell Biol. 184:639–646 10.1083/jcb.200810012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W., Kirschner M.W. 1989. Cyclin synthesis drives the early embryonic cell cycle. Nature. 339:275–280 10.1038/339275a0 [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. 1982a. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 30:675–686 10.1016/0092-8674(82)90272-0 [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. 1982b. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 30:687–696 10.1016/0092-8674(82)90273-2 [DOI] [PubMed] [Google Scholar]
- O’Farrell P.H. 2001. Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 11:512–519 10.1016/S0962-8924(01)02142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H., Stumpff J., Su T.T. 2004. Embryonic cleavage cycles: how is a mouse like a fly? Curr. Biol. 14:R35–R45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou A., McCusker D., Kellogg D.R., Sullivan W. 2008. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation. J. Cell Biol. 183:63–75 10.1083/jcb.200801153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Nurse P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 45:145–153 10.1016/0092-8674(86)90546-5 [DOI] [PubMed] [Google Scholar]
- Shermoen A.W., McCleland M.L., O’Farrell P.H. 2010. Developmental control of late replication and S phase length. Curr. Biol. 20:2067–2077 10.1016/j.cub.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon O.C., Stevenson V.A., Theurkauf W.E. 1997. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 388:93–97 10.1038/40439 [DOI] [PubMed] [Google Scholar]
- Sibon O.C., Laurençon A., Hawley R., Theurkauf W.E. 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9:302–312 10.1016/S0960-9822(99)80138-9 [DOI] [PubMed] [Google Scholar]
- Stiffler L.A., Ji J.Y., Trautmann S., Trusty C., Schubiger G. 1999. Cyclin A and B functions in the early Drosophila embryo. Development. 126:5505–5513 [DOI] [PubMed] [Google Scholar]
- Yu K.R., Saint R.B., Sullivan W. 2000. The Grapes checkpoint coordinates nuclear envelope breakdown and chromosome condensation. Nat. Cell Biol. 2:609–615 10.1038/35023555 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.