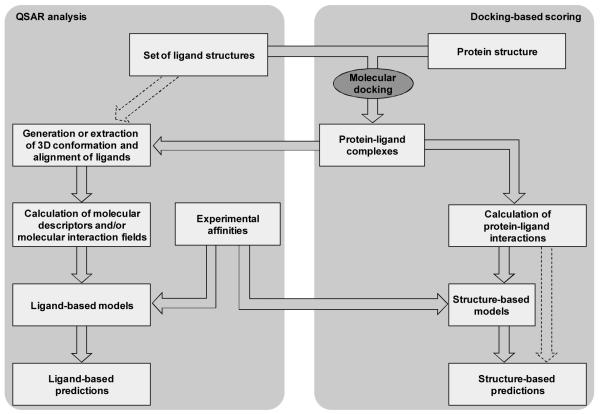
Figure 1.
Flowchart for the construction of ligand-based and structure-based models. According to this scheme, molecular docking plays a key role at the basis of both approaches. Alternatively, as indicated by the dashed arrow on the left side of the figure, a pure ligand-based approach can be adopted, in which the conformation and the alignment of the ligands are derived exclusively from their molecular features. Additionally, the scheme also illustrate that, when a training set of ligands with known activity is available, this can be used to train structure-based scoring functions. Alternatively, as indicated by the dashed arrows on the right side of the figure, a pure structure-based approach can be adopted in which prepackaged scoring functions are applied without the need for the use of a training set.