Abstract
Proteomics studies to explore global patterns of protein expression in plant and green algal systems have proliferated within the past few years. Although most of these studies have involved mapping of the proteomes of various organs, tissues, cells, or organelles, comparative proteomics experiments have also led to the identification of proteins that change in abundance in various developmental or physiological contexts. Despite the growing use of proteomics in plant studies, questions of reproducibility have not generally been addressed, nor have quantitative methods been widely used, for example, to identify protein expression classes. In this report, we use the de-etiolation (“greening”) of maize (Zea mays) chloroplasts as a model system to explore these questions, and we outline a reproducible protocol to identify changes in the plastid proteome that occur during the greening process using techniques of two-dimensional gel electrophoresis and mass spectrometry. We also evaluate hierarchical and nonhierarchical statistical methods to analyze the patterns of expression of 526 “high-quality,” unique spots on the two-dimensional gels. We conclude that Adaptive Resonance Theory 2—a nonhierarchical, neural clustering technique that has not been previously applied to gene expression data—is a powerful technique for discriminating protein expression classes during greening. Our experiments provide a foundation for the use of proteomics in the design of experiments to address fundamental questions in plant physiology and molecular biology.
Within the past few years, there have been rapid advances in proteomics technology, including the refinement of two-dimensional gel electrophoretic methods, the development of sensitive techniques of mass spectrometric protein analysis, and the acquisition of genome sequence information (Griffin and Aebersold, 2001; Mann et al., 2001). As a consequence of these developments, proteome maps and comparative proteomic studies have proliferated in plant and green algal systems. These studies have included the global mapping of proteins from maize (Zea mays) leaves (Porubleva et al., 2001), poppy (Papaver somniferum) latex (Decker et al., 2000), wheat (Triticum aestivum) grain (Skylas et al., 2001), and organs and tissues of Medicago trunculata (Watson et al., 2003). Subcellular proteomes have also been mapped, including the cell wall, plasma membrane, and endoplasmic reticulum systems from Arabidopsis (Robertson et al., 1997; Santoni et al., 1998, 2000; Prime et al., 2000; Chivasa et al., 2002), the Arabidopsis and pea (Pisum sativum) mitochondrial proteomes (Kruft et al., 2001; Millar et al., 2001; Bardel et al., 2002), lumenal and peripheral thylakoid proteins from pea chloroplasts (Peltier et al., 2000; van Wijk, 2000, 2001), lumenal proteins from Arabidopsis chloroplasts (Kieselbach et al., 2000; Schubert et al., 2002), Arabidopsis chloroplast envelope membrane proteins (Ferro et al., 2003), thylakoid membrane proteins from Chlamydomonas reinhardtii chloroplasts (Hippler et al., 2001), and plastid ribosomal subunit proteins from C. reinhardtii (Yamaguchi et al., 2002) and tobacco (Nicotiana tabacum; Yamaguchi et al., 2000; Yamaguchi and Subramanian, 2000). Comparative proteomics studies have included green versus etiolated rice (Oryza sativa) shoots (Komatsu et al., 1999), rice treated with jasmonic acid (Rakwal and Komatsu, 2000) and brassinolide (Konishi and Komatsu, 2003), Arabidopsis seed germination and priming (Gallardo et al., 2001, 2002), cell wall and extracellular matrix proteins from elicitor-treated Arabidopsis cell suspension cultures (Ndimba et al., 2003), senescing white clover (Trifolium repens; Wilson et al., 2002), and rice after mechanical wounding of the leaf sheath (Shen et al., 2003). Very few of the comparative studies have involved more than two samples (e.g. control versus treated).
One drawback to the studies to date is that questions of reproducibility generally have been treated cursorily. In addition, methods in comparative studies have frequently been qualitative in nature, and rigorous, quantitative clustering methods to identify protein expression classes have not been evaluated and exploited. In this paper, we address these questions using the light-induced de-etiolation (“greening”) of maize chloroplasts as a model experimental system. The greening of maize has long served as a model system to understand the mechanisms that regulate chloroplast biogenesis (e.g. Chen et al., 1967; Forger and Bogorad, 1973; Bogorad, 1991). Maize seeds have large energy reserves, and germinated maize seedlings can survive for several weeks in darkness. When exposed to light, photosynthetically incompetent etioplasts in dark-grown seedlings develop into photosynthetically competent chloroplasts. This involves the production of components of the photosynthetic apparatus and pronounced alterations in plastid ultrastructure that include the conversion of the distinctive prolamellar body into stromal and stacked thylakoid structures characteristic of chloroplasts (for review, see Bogorad, 1991). In maize, greening results in the formation of dimorphic mesophyll and bundle sheath cell chloroplasts that are specialized for C4 photosynthesis.
Mature chloroplasts are thought to contain about 3,000 proteins (Leister, 2003). Although metabolism in the plastid is well characterized, the functions of most of these proteins are either unknown or poorly understood. Plastid proteins are the products of both nuclear and plastid genes (for review, see Goldschmidt-Clermont, 1998). Although nuclear DNA-encoded plastid proteins are translated on 80S ribosomes and imported into the organelle posttranslationally, proteins that are products of the plastid genome are translated on 70S ribosomes, usually in a mature form. Because the plastid DNA in higher plants codes for fewer than 100 proteins, the nuclear genome is responsible for more than 95% of the different proteins in the chloroplast proteome (Martin and Herrmann, 1998). Chloroplast biogenesis is coordinated and integrated by a variety of environmental and endogenous signals, including extensive retrograde signaling between the plastid and nuclear genomes (Bogorad, 1991; Goldschmidt-Clermont, 1998; Leon et al., 1998; Bauer et al., 2001; Rodermel, 2001; Surpin et al., 2002). Although much progress has been made in deciphering these mechanisms, a more complete understanding of plastid signaling, plastid physiology, and plastid biochemistry would be facilitated by knowledge of the composition of the plastid proteome and how it changes during development.
In the present report, we use maize plastid greening as a model system to address methodological questions of reproducibility and quantification in comparative proteomics studies. As a model, maize greening offers several distinct advantages: The process has been studied, plastid metabolism is well characterized, and a formidable amount of genomics information is available for maize that facilitates spot identification on two-dimensional gels. It was our goal to develop a general protocol for comparative proteomics that could be used by a standard lab engaged in research in plant physiology and molecular biology. An assessment of issues of reproducibility and quantification and an understanding of technological limitations are a necessary prelude to the design of experiments whose goal is an understanding of fundamental mechanisms of plant biology using techniques of proteomics.
RESULTS
Experimental Design
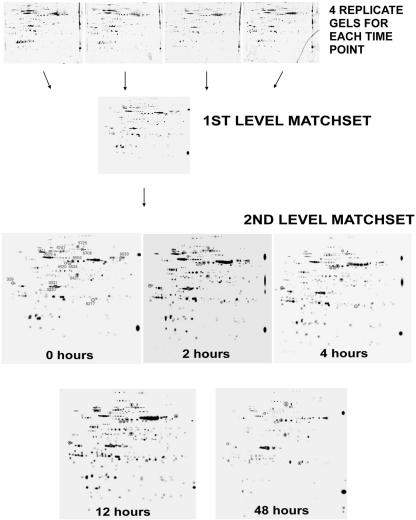
To assess changes in the maize chloroplast proteome during greening, we performed two-dimensional SDS-PAGE on proteins isolated from plastid-enriched fractions from five time points postillumination (0, 2, 4, 12, and 48 h). These times are representative of the chloroplast developmental process and were chosen based on prior work (e.g. Grebanier et al., 1979; Rodermel and Bogorad, 1985). As illustrated in Figure 1, four replicate two-dimensional gels were run for each time point. The four gels were then computationally combined into a representative standard gel, i.e. a first level match set, using PDQuest software. Although a large number of spots were included on the standard gel, only those that met several stringent criteria (classified as “high-quality” spots) were used to estimate spot quantities (see Materials and Methods). As an example, 304 different spots were included on the standard gel in the “0” hour time point, but only 271 of these were classified as high quality and subsequently used to determine protein amounts (see Table I). To compare spots from one time point to another, a second level match set was created. From this match set, the filtered spot quantities from the standard gels were assembled into a data matrix that consisted of 526 unique spots showing how each spot changed in intensity during development. Thirteen of the spots on the second level match set gels have been circled (Fig. 1) to facilitate tracking these spots in subsequent experiments.
Figure 1.
Experimental design. Plastids were isolated from five time points during maize greening (0, 2, 4, 12, and 48 h). Proteins were precipitated, and equal amounts (125 μg) were separated by electrophoresis on two-dimensional gels. Four replicate gels for each time point were computationally combined using PDQuest (Bio-Rad Laboratories, Hercules, CA) software into a first level match set (i.e. a standard gel), and quantities were determined for high-quality spots on each standard gel. A second level match set was created from the five standard gels; from this match set, a data matrix was assembled for use in subsequent statistical analyses.
Table I.
Reproducibility of two-dimensional gels
| Time | Total Spots | No. of “High-Quality” Spots | High-Quality Spots |
|---|---|---|---|
| % | |||
| 0 | 304 | 271 | 89.0 |
| 2 | 336 | 312 | 92.8 |
| 4 | 351 | 345 | 98.0 |
| 12 | 361 | 351 | 97.0 |
| 48 | 290 | 270 | 93.0 |
| Total | 1,642 | 1,549 | 94.3 |
Because the success of our experiments relied on the acquisition of a reliable, quantitative data matrix, we examined the reproducibility of our gel replicates. Visual inspection revealed that the gels were qualitatively consistent from gel-to-gel within a given time point (Fig. 1). Table I provides a quantitative measure of this by showing the fraction of spots on each of the standard gels (first level match set) that were classified as high quality. Using the example above, 89% of the spots on the standard gel in the “0” hour time point were considered to be high quality (i.e. 271 of 304 total spots). Overall, the data reveal that nearly 95% of the 1,642 spots on our gels were high quality, suggesting excellent reproducibility. Of the 1,549 high-quality spots, 526 were unique and were used in the analyses described below, i.e. some of the 526 proteins were detectable at all five time points, whereas others were not.
Protein Identification
Of the most intense 526 high-quality spots, 401 were excised from the two-dimensional gels, trypsin digested, and analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (see “Materials and Methods”). Good spectra were obtained from 166 of the digests (41.4%). Using Protein Prospector software (University of California, San Francisco), the peptide mass fingerprints from these spectra were compared with translation products from expressed sequence tag and genomic DNA sequence databases that had been theoretically digested with trypsin. Because this software requires that each fingerprint be searched individually, we developed a program to facilitate this process (available at http://baker1.zool.iastate.edu/batch_msfit.html). This program interacts with Protein Prospector and submits peptide mass fingerprints in batch mode for database comparison.
Of the 166 spectra, 93.4% returned an identification match. Using stringent criteria (see “Materials and Methods”), we were able to identify 54 of the spots unambiguously (Table II). The theoretical and experimental masses and pIs matched closely for 47 of the 54 spots, but for seven spots, the theoretical and experimental masses, but not pIs, approximately matched (Table II, see footnote a). For instance, inosine monophosphate dehydrogenase is predicted to have a molecular mass of 11,784 D, as observed on the two-dimensional gels, but its predicted pI (9.78) is much higher than is seen on the gels (less than 7). This seeming discrepancy might be a consequence of posttranslational modification (Battey et al., 1993). From Table II, it is clear that some of the 54 proteins are represented by more than one spot. These spots might be isozymes or posttranslational modifications of a single protein. Yet, because many of these proteins are coded for by single genes on the plastid genome (for example, atpA and atpB, for the α- and β-subunits of the proton ATP synthase), it is likely that, at least in these cases, the multiple spots represent posttranslational modifications. One feature of Table II is that the predicted molecular masses of the multiple forms of a given protein differ, e.g. six different molecular masses are predicted for the 11 ATP synthase β-subunit spots. The major reason for this is that the peptide fragment patterns from the 11 spots matched fragments of plastid atpB genes from different species in the databases; among plastid genes, atpB is moderately conserved among higher plants (Rodermel and Bogorad, 1987).
Table II.
Mass spectrometric identification of spots on two-dimensional gels
| Spot No. | Database | Gene Identifier | Annotation | MOWSE (Molecular Weight Search) Score | Peptides Matched (Total No. of Peptides) | Predicted Molecular Mass (D) | Predicted pl | Coveragea | Codine Site | Plastid Targetingb |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| 3317 | National Center for Biotechnology Information (NCBI) | 134102 | 60-kD chaperonin alpha subunit (Cpn60), chloroplast precursor | 5.42 E + 03 | 7 (32) | 57,521 | 4.83 | 13 | Nucleus | Yes |
| 3603 | NCBI | 134102 | 60-kD chaperonin alpha subunit (Cpn60), chloroplast precursor | 5.42 E+03 | 7 (32) | 57,521 | 4.83 | 13 | Nucleus | Yes |
| 3610 | NCBI | 134102 | 60-kD chaperonin alpha subunit (Cpn60), chloroplast precursor | 1.00 E+04 | 7 (22) | 57,521 | 4.83 | 16 | Nucleus | Yes |
| 3615 | NCBI | 134102 | 60-kD chaperonin alpha subunit (Cpn60), chloroplast precursor | 1.60 E+05 | 9 (31) | 57,521 | 4.83 | 18 | Nucleus | Yes |
| 3324 | The Institute for Genomic Research (TIGR) Zm | TC88712 | 20-kD chaperonin (Cpn20), chloroplast precursor | 9.88 E+5 | 8 (25) | 27,095 | 6.25 | 37 | Nucleus | Yes |
| 4209 | TIGR Zm | TC88576* | 20-kD chaperonin (Cpn20), chloroplast precursor | 1.02 E+7 | 8 (19) | 26,569 | 8.48 | 45 | Nucleus | Yes |
| 3320 | NCBI | 15912247 or 13926292 | 33-kD subunit of OEC, PSI | 93.4 | 4 (22) | 35,128 | 5.55 | 11 | Nucleus | Yes |
| 3330 | NCBI | 15408655 | 33-kD subunit of OEC, PSI (pulative) | 984 | 4 (25) | 34,861 | 6.09 | 17 | Nucleus | Yes |
| 4305 | TIGR Zm | TC81083 | 33-kD subunit of OEC, PSII | 2.13 E+6 | 9 (26) | 37,531 | 5.92 | 43 | Nucleus | Yes |
| 6537 | TIGR Zm | TC88557 | Acetyl CoA carboxylase | 214 | 5 (22) | 258,314 | 5.93 | 2 | Nucleus | Yes |
| 7537 | NCBI | 7438099 | Acetyl CoA carboxylase | 1.25 E+3 | 9 (32) | 252,131 | 5.91 | 6 | Nucleus | No |
| 7616 | TIGR Zm | TC90687 | ATPase subunit alpha | 5.48 E+5 | 8 (31) | 55,707 | 5.86 | 23 | Plastid | N.A.c |
| 8632 | TIGR Zm | TC90687 | ATPase subunit alpha | 1.36 E+5 | 8 (26) | 55,707 | 5.86 | 19 | Plastid | N.A. |
| 8633 | TIGR Zm | TC90687 | ATPase subunit alpha | 2.87 E+5 | 7 (30) | 55,707 | 5.86 | 19 | Plastid | N.A. |
| 1246 | TIGR Zm | TC85581 | ATPase subunit beta | 1.69 E+7 | 11 (29) | 54,041 | 5.31 | 28 | Plastid | N.A. |
| 4520 | NCBI | 629818 | ATPase subunit beta | 7.15 E+4 | 7 (16) | 59,249 | 5.56 | 18 | Plastid | N.A. |
| 4533 | NCBI | 552857 | ATPase subunit beta | 5.82 E+5 | 9 (23) | 53,954 | 5.30 | 19 | Plastid | N.A. |
| 4534 | NCBI | 629818 | ATPase subunit beta | 1.54 E+5 | 7 (14) | 59,249 | 5.56 | 20 | Plastid | N.A. |
| 5504 | NCBI | 552857 | ATPase subunit beta | 3.52 E+3 | 5 (24) | 53,954 | 5.30 | 17 | Plastid | N.A. |
| 5507 | NCBI | 629818 | ATPase subunit beta | 7.23 E3 | 6 (22) | 59,249 | 5.56 | 14 | Plastid | N.A. |
| 5517 | NCBI | 6815115 | ATPase subunit beta | 4.63 E+5 | 11 (30) | 53,997 | 5.38 | 23 | Plastid | N.A. |
| 5518 | NCBI | 629818 | ATPase subunit beta | 1.14 E+3 | 5 (19) | 59,149 | 5.56 | 11 | Plastid | N.A. |
| 5725 | NCBI | 874823 | ATPase subunit beta | 4.86 E+4 | 7 (32) | 53,717 | 5.20 | 18 | Plastid | N.A. |
| 6511 | NCBI | 552857 | ATPase subunit beta | 2.54 E+5 | 9 (19) | 53,954 | 5.30 | 24 | Plastid | N.A. |
| 6708 | TIGR Zm | TC85581 | ATPase subunit beta | 3.69 E+6 | 12 (28) | 54,041 | 5.31 | 30 | Plastid | N.A. |
| 201 | TIGR Zm | TC86875 | ATPase subunit delta | 1.22 E+4 | 5 (17) | 20,206 | 4.35 | 38 | Nucleus | Yes |
| 6439 | TIGR Zm | TC88574 | ATPase subunit gamma | 104 | 4 (17) | 42,529 | 8.80 | 11 | Nucleus | Yes |
| 7414 | TIGR Zm | TC88574 | ATPase subunit gamma | 1.21 E+3 | 7 (14) | 42,529 | 8.83 | 18 | Nucleus | Yes |
| 8520 | NCBI | 7489718 | Beta-D-glucosidase, glu2 precursor | 1.74 E+2 | 7 (16) | 64,112 | 6.72 | 16 | Nucleus | Yes |
| % | ||||||||||
| 5406 | TIGR Zm | TC85027 | Chloroplast NADP-malate dehydrogenase | 9.64E+3 | 6 (30) | 49,481 | 6.62 | 17 | Nucleus | Yes |
| 6705 | NCBI | 5360574 | ClpC protease | 5.16E+3 | 6 (19) | 105,740 | 6.06 | 8 | Nucleus | Yes |
| 6711 | NCBI | 2921158 | ClpC protease | 3.95E+7 | 14 (26) | 103,456 | 6.27 | 14 | Nucleus | Yes |
| 3632 | NCBI | 16444957 | Cryptochrome 1 | 1.05E+3 | 5 (20) | 79,300 | 5.37 | 9 | Nucleus | No |
| 4738 | NCBI | 16444957 | Cryptochrome 1 | 631 | 4 (26) | 79,301 | 5.37 | 8 | Nucleus | No |
| 4741 | NCBI | 16444957 | Cryptochrome 1 | 2.63E+3 | 5 (32) | 79,300 | 5.37 | 10 | Nucleus | No |
| 4417 | NCBI | 15809970 | Enolase (2 phospho D glycerate hydroylase) | 1.47E+3 | 5 (31) | 47,704 | 5.74 | 18 | Nucleus | No |
| 2714 | NCBI | 6746592 | Hsp70 | 9.32E+3 | 7 (33) | 77,106 | 5.13 | 10 | Nucleus | Yes |
| 2720 | NCBI | 6746592 | Hsp70 | 5.55E+4 | 8 (27) | 77,106 | 5.13 | 11 | Nucleus | Yes |
| 329 | NCBI | 100903 | Nucleic acid-binding protein (NABP) | 1.51E+3 | 4 (24) | 33,117 | 4.60 | 18 | Nucleus | Yes |
| 4409 | TIGR Zm | TC82882 | Phosphoglycerate kinase, chloroplast precursor | 2.44E+4 | 5 (28) | 24,114 | 9.12 | 33 | Nucleus | Yes |
| 4219 | TIGR Zm | TC89102 | Plastid-specific ribosomal protein “2” | 1.08E+5 | 8 (17) | 26,232 | 8.46 | 25 | Nucleus | Yes |
| 5421 | NCBI | 13489165 | Beta amylase | 3.37E+3 | 8 (32) | 57,953 | 6.74 | 19 | Nucleus | Yes |
| 6441 | TIGR Zm | TC82049 | SU1 isoamylase | 468 | 4 (21) | 91,619 | 5.85 | 6 | Nucleus | Yes |
| 7313 | TIGR Zm | TC81138 | Sucrose synthase (UDP-glucose: D fructose-2-glucosyl-transferase) | 152 | 4 (9) | 93,696 | 6.18 | 4 | Nucleus | No |
| 6402 | TIGR Zm | TC84809 | Glyceraldehyde-3-phosphate dehydrogenase | 3.62E+4 | 5 (25) | 46,952 | 5.95 | 18 | Nucleus | Yes |
| 3629 | NCB1 | 12325133 | Unknown protein | 193 | 6 (25) | 66,659 | 5.06 | 12 | Nucleus | No |
| 7230 | NCB1 | 16905193 | Hypothetical protein | 3.09E+2 | 4 (23) | 26,072 | 6.34 | 21 | Nucleus | No |
| 1353 | NCBI | 7459088 | Hypothetical protein | 4.05E+3 | 4 (37) | 11,782 | 4.94d | 28 | Nucleus | No |
| 3331 | NCBI | 16604341 | Unknown protein | 2.36E+3 | 5 (20) | 39,424 | 5.12d | 13 | Nucleus | No |
| 1145 | TIGR Zm | TC81772 | Inosinc monophosphate dehydrogenase | 3.65E+4 | 6 (26) | 11,784 | 9.78d | 61 | Nucleus | No |
| 2528 | NCBI | 3915111 | Cytochrome P450 (C4H) | 1.5 E+3 | 5 (23) | 58,011 | 9.05d | 16 | Nucleus | No |
| 3602 | NCBI | 15554512 | Male sterility 1 protein | 1.40 E14 | 7 (32) | 76,974.2 | 7.82d | 17 | Nucleus | No |
| 4301 | TIGR Zm | TC89587 | Mitotic spindle checkpoint protein MAD2 | 2.61 E4 | 4 (21) | 26,664 | 4.73d | 24 | Nucleus | No |
| 6217 | TIGR Zm | TC89879 | Putative protein phosphatase | 1.03E+3 | 4 (37) | 25,946 | 10.38d | 25 | Nucleus | No |
Percent of predicted protein sequence the peptides matched. b Plastid targeting sequence determined by ChloroP software. c N.A., Not applicable, protein is plastid encoded. d Predicted pl does not match the pl of the spot on the gel.
Taking into account the multiplicity of spots, we were able to identify a total of 26 unique proteins on our gels. These proteins fall into several predominant classes. Proteins that are involved in the light reactions of photosynthesis include four of the five subunits of the extrinsic CF1 complex of the proton ATP synthase (Groth and Strotmann, 1999) and the 33-kD subunit of the oxygen-evolving complex of PSII (Liveanu et al., 1986; Hankamer et al., 1997). The photosynthetic carbon assimilation cycle is represented by β-amylase, isoamlylase, glyceraldehyde-3-phosphate dehydrogenase, NADP-malate dehydrogenase, phosphoglycerate kinase, and Suc synthase. Chaperones include the α-subunit of chaperonin 60, originally called the Rubisco subunit-binding protein (Hemmingsen et al., 1988; Martel et al., 1990); chaperonin 20, a regulator of chaperonin-mediated protein folding (Koumoto et al., 2001)); and Hsp70, a member of the versatile class of 70-kD heat shock proteins that mediate protein transport, folding, and assembly (Strzalka et al., 1994; Drzymalla et al., 1996; Sung et al., 2001). We also identified ClpC, the ATPase (regulatory) subunit of the ClpC Ser-type stromal protease that also serves as a chaperone (Ostersetzer and Adam, 1996; Nielsen et al., 1997; Adam et al., 2001). Other enzymes involved in plastid metabolism include acetyl-coA carboxylase, which mediates the synthesis of malonyl-coA during fatty acid biosynthesis (Ke et al., 2000); β-d-glucosidase, involved in the hydrolysis of many plastid compounds (Esen, 1992); nucleic acid-binding protein, likely involved in posttranscriptional control of plastid gene expression (Cook and Walker, 1992); and ribosomal protein “2,” a stromal RNA-binding protein that might be a component of the plastid ribosomal 30S subunit (K. Yamaguchi and A.R. Subramanian, unpublished data).
Nine of the proteins we were able to identify unambiguously on our gels did not localize to the chloroplast using the transit peptide prediction software ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). Other prediction programs, such as TargetP and Predotar, gave similar results. In addition to Suc synthase, mentioned above, these proteins included cryptochrome 1, a blue light photoreceptor (Christie and Briggs, 2001); cinnamate-4-hydroxylase, the first cytochrome P450-dependent monooxygenase of the phenylpropanoid pathway (Bell-Lelong et al., 1997); enolase, a glycolytic enzyme; inosine monophosphate dehydrogenase, involved in nucleotide catabolism; the cytosolic form of acetyl-coA carboxylase, which is used in fatty acid elongation and flavonoid synthesis (Roesler et al., 1994); and uncharacterized proteins that have been annotated as “male sterility 1 protein,” “mitotic spindle checkpoint protein,” and “protein phosphatase.” In addition, we identified four “unknown” or “hypothetical” proteins, none of which were predicted to have plastid transit peptides.
Expression Patterns during Plastid Biogenesis
Early one-dimensional SDS-PAGE analyses were able to distinguish three major patterns of change in plastid proteins during maize greening: an increasing trend, a decreasing trend, and no change (Grebanier et al., 1979). Consistent with these patterns, preliminary principal components analysis (PCA; Jolliffe, 1986) of our data showed that general increases and decreases in protein abundance accounted for about 49% of the variability in the data set (data not shown). To examine our data set in greater detail, we employed three clustering techniques. Clustering techniques generally fall into two broad categories: hierarchical and nonhierarchical. We first tried a hierarchical method, pair-wise average linkage (PAL). PAL operates by defining two entities, here protein spots, as similar and then reiteratively adds other similar entities resulting in a tree-like diagram. Each “leaf” on the tree represents a unit (i.e. a spot); in principle, the branches represent clusters of spots with similar expression patterns.
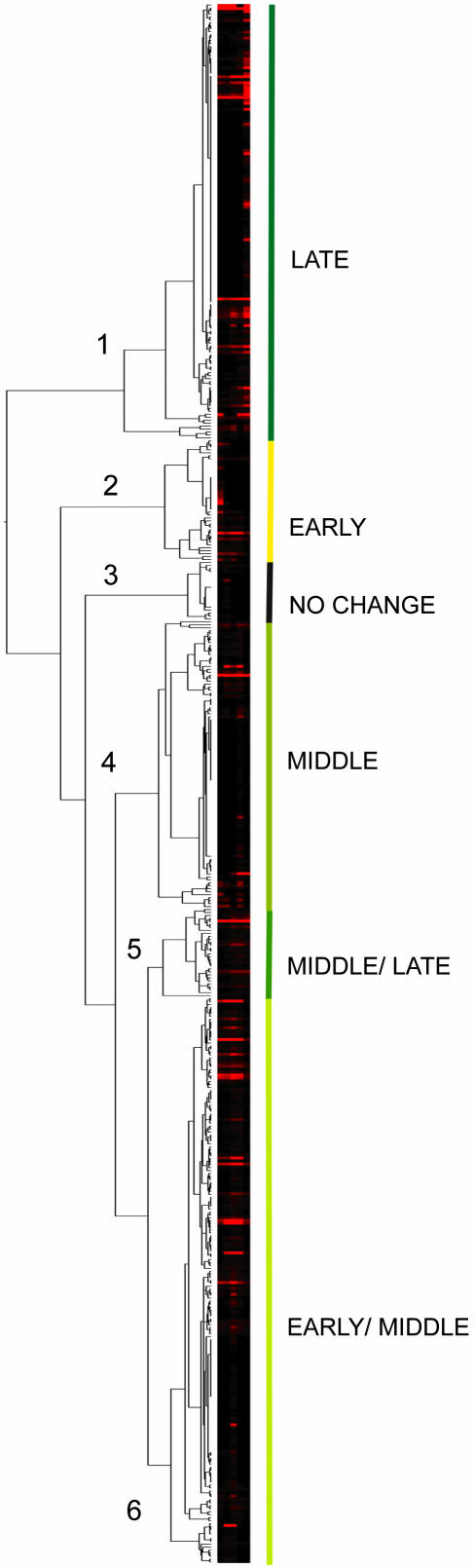
As illustrated in Figure 2, PAL analysis of our data gave rise to a tree that can be divided into six main branches. The 526 “leaves” on this tree correspond to the 526 proteins whose patterns of expression we were able to track during the greening process. An examination of this tree reveals that there is a lack of uniform expression within each branch, a problem previously pointed out by others in expression data analyses (Sherlock, 2000). This might be because of the relatively small size of our data set: Trees obtained by hierarchical methods are greatly influenced by the early decisions, and if the early clusters portray inaccurate relationships, then the tree can be misleading (Dopazo et al., 2001). Despite the lack of uniform expression within each branch, we classified the six branches according to the predominant mode of expression of the proteins in each branch. Proteins in the “early” branch are, in general, abundantly expressed at 0, 2, or 4 h, but not at other time points; spots in the “middle” branch have high expression at 12 h but not at other time points; spots in the “late” branch have high expression at 48 h but not at the other times; “early/middle” and “middle/late” describe branches whose spots fall into two expression categories; and the “no change” branch describes proteins for which no obvious pattern is evident. Considering the size of each branch, these data suggest that a preponderance of the 526 protein spots are expressed early in chloroplast biogenesis, whereas fewer are expressed late in development.
Figure 2.
Hierarchical PAL clustering. Results are displayed as a tree with leaves representing 526 individual protein spots and branches as clusters. The level of expression for each protein ranges from high (red) to low (black). The general trends of expression of the spots within each of the six main branches are classified as early (high expression at 0, 2, or 4 h), middle (high expression at 12 h), or late (high expression at 48 h), or combinations thereof.
Next, we used nonhierarchical clustering techniques to analyze our data. Nonhierarchical clustering does not define relationships between clusters; rather, it defines a set of clusters and then partitions entities to those clusters while minimizing the within-cluster dispersion. The first nonhierarchical clustering method we used was Adaptive Resonance Theory 2 (ART2; Carpenter et al., 1991), a method that has not been applied previously to gene expression data but has been used in other fields such as microgravity (Smith and Sinha, 1999) and image classification (Hadjiiski et al., 1999). ART2 is an unsupervised neural network that mimics connections between neurons. It collapses the dimensionality of the data and defines a number of clusters (cells) using a vigilance value. It begins by normalizing the data, then chooses two data points and calculates their similarity. If the similarity value exceeds the vigilance value, a cluster is created consisting of the similar data points. If the similarity between the two data points is below the vigilance value, a new cluster is created. This process repeats reiteratively, resulting in a grid in which each cell shows an expression pattern representative of that cluster. Varying the vigilance value (between 0 and 1) causes the number of clusters to change. The higher the vigilance value, the more sensitive the network is to dissimilarities in patterns. Therefore, high vigilance values will result in more categories.
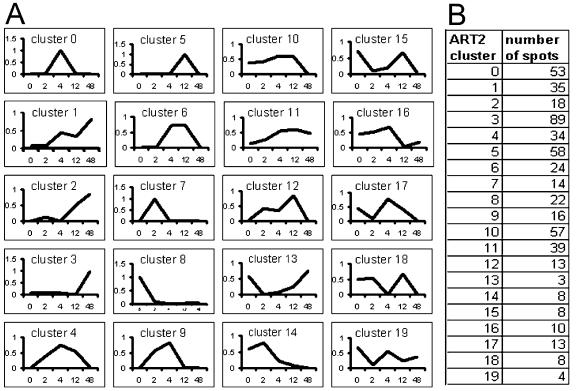
To implement the ART2 algorithm, we wrote software based on the method described by Gallant (1993) to analyze normalized medians. Four parameters (α, β, θ, and ρ) are necessary for this analysis and were set at: α (similarity parameter) = (0.5/SQRT(N)), β (update parameter) = (0.5/SQRT(N)), θ (normalization parameter) = (0.15), and ρ (vigilance) = (0.85), where n = the total number of spots in the data set (526). Preliminary experiments using yeast (Saccharomyces cerevisiae) microarray data revealed that a good range for the vigilance value is between 0.8 and 0.95: A vigilance value less than 0.8 results in categories that are too broad, whereas a value greater than 0.95 results in too many categories (X. Zhang and V. Honavar, unpublished data). Figure 3 shows the clustering results of our data using a vigilance value of 0.85. Using this value, the expression patterns were divided into 20 clusters, numbered 0 through 19. Consistent with the early data of Grebanier et al. (1979), about 35% of the spots showed a general increase during chloroplast biogenesis (clusters 1–3 and 11), whereas 17% showed a general decrease (clusters 8, 10, and 14). However, the remainder of the proteins have more complicated patterns of increase and decrease.
Figure 3.
Nonhierarchical ART2 clustering. A, Representative expression patterns for each of the 20 clusters obtained using ART2 analysis; the x axis represents time point during greening, and the y axis represents the normalized protein amount. B, Cluster number and number of spots within each cluster.
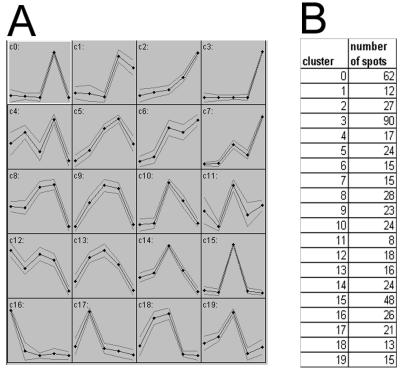
In addition to ART2, we used another nonhierarchical neural network clustering method, self-organized mapping (SOM), to analyze our data. SOM has been used previously for microarray data (e.g. Maleck et al., 2000; Chen et al., 2002) but not for proteomics data. SOM clustering works well for large data sets because neural networks are less influenced by noise and the shape of the data distribution (Dopazo et al., 2001). The SOM algorithm maps high-dimensional data onto an ordered two-dimensional space, resulting in an ordered grid where each cell represents a model pattern for the corresponding set of data points. For expression data, the pattern inside each cell represents the expression pattern over time for that cluster. Cells that have similar patterns are closer to one another within the grid. However, there are two disadvantages to the SOM method: (a) The user must arbitrarily predefine the number of clusters, and (b) noisy data patterns are partitioned into existing clusters instead of being separated from stronger patterns. Figure 4 shows the results of SOM analysis of our data using a cluster number of 20. This number was chosen to facilitate a comparison of the SOM and ART2 methods. We obtained very similar clusters with the SOM and ART2 methods, with approximately 30% of the spots showing a general increase during greening (clusters 1–3, 6, and 7) and about 18% showing a general decrease (clusters 8, 12, 14, and 16). As with the ART2 analysis, approximately 50% of the spots showed more complex patterns of expression.
Figure 4.
Nonhierarchical SOM clustering. A, Representative expression patterns from SOM analysis. Thick line, Expression pattern; Thin lines, sd; x axis, “time point during greening”; y axis, represents normalized protein amount. B, Cluster number and number of spots within each cluster.
DISCUSSION
Plant proteomic studies published to date have focused on mapping of the proteomes of various organs, tissues, and cellular components, or on comparing protein differences between two or more samples (see above). However, quantitative measures of reproducibility were not reported in these studies, nor were rigorous quantitative analyses conducted to group proteins into expression classes (e.g. clustering analyses). As examples of methodologies involving comparisons of more than two samples, two recent studies have investigated temporal changes in plant proteomes involving up to four different time points (Wilson et al., 2002; Shen et al., 2003). Wilson et al. (2002) examined changes in the proteomes of “enriched chloroplast” fractions of senescing white clover. Proteins were isolated from individual leaves of “mature green,” “early senescent,” and “late senescent” plants, and 590 spots were resolved on the gels. The quantity of each protein was assessed as a percentage of the total amount of staining on each gel, and of the 590 spots, it was possible to qualitatively assign about 50% to four expression groups based on their patterns of change during leaf ontogeny. However, high errors were obtained for the relative staining intensities (abundances) of many spots, suggesting that there was high variability between the different gels at each time point because of developmental and/or technical factors. Of the 590 spots, only six plastid proteins could be firmly identified, illustrating (at least in part) the difficulties of performing proteomics with an organism for which limited genomics information is available. Although the data in these experiments are useful in providing descriptive information about groups of proteins that change in abundance in a coordinate fashion during leaf development, the assignment of proteins to a given class is rather arbitrary because quantitative clustering analyses were not performed.
In another “timed series” experiment, Shen et al. (2003) explored changes in the rice proteome at four time points (from 0–48 h) after mechanical wounding of the leaf sheath. Soluble rice leaf proteins were isolated, and about 400 spots were resolved on two-dimensional gels. Of these spots, 29 were qualitatively observed to change in abundance after wounding (19 were “up-regulated” and 10 were “down-regulated”). Although clustering analyses were not performed to assign proteins to a given class, there appeared to be good gel reproducibility because there was little variability in staining intensity on replicate gels from each time point. Even though the rice genome has been sequenced, these authors were able to identify only 14 of the 29 spots by MALDI-TOF and/or Edman sequencing. Only 10 of these spots represented unique proteins.
The data in this paper provide a reliable method to assess patterns of change in the plastid proteome during development. Using our methodology, we were able to obtain reproducible, replicate gels and to classify nearly 95% of the visible spots on these gels as high quality, facilitating estimations of spot quantities (protein amounts). As other researchers have noted (e.g. Porubleva et al., 2001), a major stumbling block in plant proteomic projects is the lack of reliable means of spot identification. There are three levels at which a firm identification can be compromised: (a) Once a spot is digested, a good spectrum cannot be obtained by mass spectrometry; (b) for spots with good spectra, database searches for possible identification yield no matches; and (c) for those spots with possible identifications, the identifications are tenuous until supported by experimental data. Impediments at any of these steps cause the final number of spots that can be identified with confidence to be low. Under the strict criteria applied in the present study, only 13.5% (54/401) of the original trypsin-digested samples could be identified with certainty. Although we obtained tentative classifications for another 25%, we could not confirm these spots unambiguously.
Of the 54 high-confidence spots, most are bona fide plastid proteins. Yet, some “non-plastid” proteins were also found. This might not be surprising because we used only crude organelle preparations for our two-dimensional gels. On the other hand, not all plastid proteins have targeting sequences (Schleiff and Soll, 2000); in addition, chloroplast-targeting algorithms are not always good at predicting these sequences. Thus, some of the proteins we classified as “non-plastid” might in fact be bona fide plastid proteins. Further experiments are necessary to determine the location of these proteins.
The “non-plastid” protein class included four “unknown” or “hypothetical” proteins. Similarity searches to known protein motifs or domains did not yield clues as to the function of these proteins. However, protein threading using the software LOOPP (http://ser-loopp.tc.cornell.edu/loopp.html) gave several high-confidence matches for one of the “unknowns” (spot 3331). LOOPP predicts protein function based on amino acid sequence-to-sequence, sequence-to-protein structure, and structure-to-structure similarity. Using this program, spot 3331 showed similarity to three different proteins. The highest was to an Escherichia coli Leu/Ile/Val-binding protein [Protein Data Bank (PDB) identifier 2liv] that interacts with a set of membrane proteins to transport branched chain amino acids into the cytoplasm (Landick and Oxender, 1985). The next highest similarity was to collagenase (PDB identifier1fbl), which is a member of a family of zinc-dependent matrix metalloproteases (Li et al., 1995). The lowest similarity was to the E. coli matrix porin outer membrane protein F (PDB identifier 1bt9). Further studies are necessary to determine whether protein 3331 has any of these functions.
Not surprisingly, all of the proteins we were able to identify with confidence are soluble or peripheral membrane proteins, most likely because integral membrane proteins are difficult to resolve using standard isoelectric focusing (IEF) and two-dimensional gel procedures (Molloy, 2000). However, we could not identify some prominent soluble stromal proteins on our gels, such as Rubisco and phosphoenolpyruvate carboxylase. Similar results were reported by Porubleva et al. (2001) in their mapping studies of the total leaf maize proteome. The lack of Rubisco, which is located in bundle sheath cell chloroplasts, might be because of a higher abundance of mesophyll cells than bundle sheath cells in our cell fractionations (Sheen and Bogorad, 1985), whereas the absence of phosphoenolpyruvate carboxylase (109 kD) might be because of a general underrepresentation of high-molecular mass proteins on two-dimensional gels.
Clustering Analyses
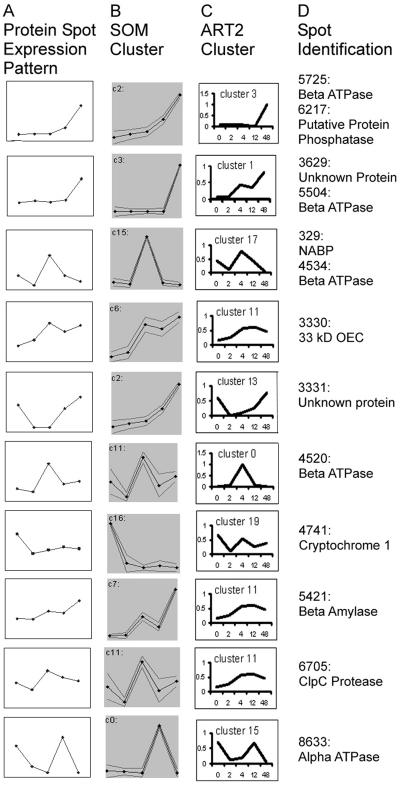
Although a growing number of comparative proteomics studies have been reported in plant systems (see above), the grouping of proteins into expression classes has generally been qualitative, and rigorous quantitative measures have been lacking. In this paper, we evaluated three types of clustering approaches to determine patterns of change in protein expression using a developmental sequence (greening) as a model system. We found that nonhierarchical neural network clustering methods are superior to hierarchical techniques, given the size of our data set. Of these, ART2 is preferable to SOM because it eliminates the need for the user to predefine the number of clusters. However, the user still needs to define the vigilance value. Figure 5 shows expression profiles of 13 representative proteins of the 54 total in Table II and the clusters into which these proteins were assigned by the ART2 and SOM methods. The expression profiles were derived from the standard gels of the five time points. In most cases, the protein profiles closely match the patterns of both clusters, but there are exceptions, e.g. spot 3331 (an “unknown” protein), which more closely matches the profile of ART2 cluster 13 than SOM cluster 2. Yet, such exceptions are rare, and we conclude that both ART2 and SOM provide an accurate reflection of the actual patterns of change that occur in individual proteins.
Figure 5.
Comparison of ART2 and SOM clustering methods. A, Protein spot expression patterns from the standard gels for 13 representative proteins from Table II; the x axis is “time point during greening” and the y axis is spot quantity. B, Pattern of SOM assigned cluster for protein. C, Pattern of ART2 assigned cluster for protein. D, Spot identification of each protein (same as the circled spots in Fig. 1).
The ART2 clusters into which the 54 proteins in Tables II and III fall have been included in Table III. Several trends emerge from the data. One is that members of a given functional class of protein are generally coordinately regulated in expression, at least during part of plastid development. For instance, the enzymes of photosynthetic carbon assimilation generally increase during early development and then reach a plateau (e.g. β-amylase, NADP-malate dehydrogenase, and phosphoglycerate kinase), continue to increase (e.g. glyceraldehyde-3-phosphate dehydrogenase), or decrease (e.g. isoamylase). The phosphoglycerate kinase pattern resembles that of the mRNA expression profile of PGK (the gene for phosphoglycerate kinase) in greening tobacco (Bringloe et al., 1996), and the pattern of glyceraldehyde-3-phosphate dehydrogenase expression is similar to the mRNA expression profiles of both genes for this enzyme (GapA and GapB) after the illumination of mature, dark-adapted Arabidopsis (Dewdney et al., 1993). Although early increases in expression of photosynthetic carbon assimilation proteins might not be surprising because the plastid is assembling the machinery for photosynthesis during this time, the lack of a single expression pattern for these proteins perhaps was presaged by early experiments in which the in vitro activities of several Calvin cycle enzymes were monitored during the greening process (Chen et al., 1967).
Table III.
Classes of proteins identified on two-dimensional gels and their patterns of expression during maize greening
| Category | Spot No. | ART2 Classification |
|---|---|---|
| Light reactions of photosynthesis | ||
| ATPase alpha subunit | 8632, 7616, 8633 | 1, 12, 15 |
| ATPase beta subunit | 4520, 4533, 5504, 5517, 5725, 6511, 6708, 5507, 1246, 5518, 4534 | 0, 1, 1, 3, 3, 3, 3, 6, 10, 10, 17 |
| ATPase delta subunit | 201 | 1 |
| ATPase gamma subunit | 6439, 7414 | 1, 1 |
| 33 kD OEC PSII | 3320, 3330, 4305 | 3, 11, 18 |
| Photosynthetic carbon assimilation | ||
| Beta amylase | 5421 | 11 |
| Glyceraldehyde-3-phosphate dehydrogenase | 6402 | 1 |
| Isomylase | 6441 | 5 |
| NADP-malate dehydrogenase | 5406 | 11 |
| Phosphoglycerate kinase | 4409 | 11 |
| Plastid chaperones, proteases | ||
| Cpn60 (alpha subunit) | 3603, 3610, 3615, 3317 | 10, 10, 10, 18 |
| Cpn20 | 3324, 4209 | 4, 10 |
| ClpC | 6705, 6711 | 11, 11 |
| Hsp70 | 2714, 2720 | 11, 11 |
| Plastid metabolism (miscellaneous) | ||
| Acetyl-coA carboxylase | 6537 | 10 |
| Beta-D-glucosidase | 8520 | 10 |
| Nucleic acid-binding protein (NABP) | 329 | 17 |
| Ribosomal protein “2” | 4219 | 10 |
| Non-plastid | ||
| Acetyl-coA-carboxylase | 7537 | 4 |
| Cryptochrome 1 | 3632, 4738, 4741 | 3, 8, 19 |
| Cytochrome P450-dependent C4H | 2528 | 10 |
| Enolase | 4417 | 10 |
| Inosine monophosphate dehydrogenase | 1145 | 1 |
| Male sterility 1 protein | 3602 | 10 |
| Mitotic spindle checkpoint protein | 4301 | 3 |
| Sucrose synthase | 7313 | 4 |
| Protein phosphatase | 6217 | 3 |
| Unknown, Hypothetical | 3629 | 1 |
| 3331 | 13 | |
| 7230 | 2 | |
| 1353 | 10 |
The most abundant proteins on our gels were the α-, β-, δ-, and γ-subunits of the proton ATPase. Because the α- and β-subunits are coded for by single-copy plastid genes, it is likely that the multiple spots for these proteins arise as a consequence of posttranslational modifications. This might also explain why there are at least two spots for the γ-subunit. Yet, because the γ-subunits are coded for by nuclear genes, it cannot be ruled out that these spots represent isozymes. The expression patterns of all four ATPase subunits fall into cluster 1 (a continual increase during greening), but some of the spots for the α- and β-subunits also fall into additional clusters. This suggests that different forms of these proteins function during specific times during light-induced chloroplast differentiation. Early studies by de Heij et al. (1984) showed that the α-, β-, γ-, and ε-subunits of the plastid proton ATPase increase 10-fold during the greening of duckweed (Spirodela oligorhiza), as measured by western-blot analysis. If duckweed resembles maize, it is likely that the general increases in protein expression in duckweed reflect a summation of the patterns of change of all the different forms of each subunit, masking underlying differences.
Table II shows that in addition to the ATPase subunits, multiple spots are represented by the α-subunit of the 60-kD and 20-kD chaperonins. These proteins both fall into cluster 10, having a moderate increase during most of development and a decrease at 48 h. A similar pattern has been observed for the chloroplast 60-kD chaperonin during the deetiolation of pumpkin (Cucurbita pepo; Strzalka et al., 1994). However, some isoforms of the 60- and 20-kD proteins have more complex patterns of change, again consistent with the notion that different enzyme forms might be required at discrete times during development.
Although one can vary the cluster number in ART2 by varying the vigilance value, our results are consistent with the idea that there is a wider range of patterns of change in protein expression during the greening process than reported in the first proteomic studies of this process using one-dimensional gels nearly 25 years ago, in which three expression classes were identified (Grebanier et al., 1979). The significance of these patterns is unclear, but to gain insight into the responsible mechanisms, we are using techniques of proteomics to analyze mutants that are perturbed in the process of chloroplast development. Future experiments will also be directed toward identifying more chloroplast proteins on our two-dimensional gels, both to amplify our understanding of how chloroplast development is controlled during greening and to identify unknown proteins that might be important regulators of this process. Although spot identification should improve as the maize genomic becomes more complete, spot identification could also be enhanced by using techniques such as Edman degradation, tandem mass spectrometry, or isotope-coded affinity tags (Gygi et al., 1999; Hubbard, 2002). Advances in methods of sample preparation and IEF should also improve the efficiency of proteome analysis for proteins previously intractable to two-dimensional gel analysis because of their low abundance, poor solubility, or high basicity (Rabilloud et al., 1999; Görg et al., 2000; Herbert and Righetti, 2000).
In conclusion, using the greening of maize chloroplasts as a model system, we developed a general protocol that can be used to generate high-quality, reproducible data sets for comparative plant proteomics. We also evaluated quantitative procedures that can be used to group proteins from these data sets into expression classes and showed that ART2 provides reliable clusters. Importantly, our procedures can be employed by a standard research lab that is interested in functional genomics to probe the function of a protein of interest, for example, by comparing the proteomes of wild-type and knockout mutants.
MATERIALS AND METHODS
Plant Growth
Maize (Zea mays) kernels were soaked overnight in water, planted in a mixture of 50% (w/v) peat moss, 40% (w/v) perlite, and 10% (w/v) mineral soil in 6-inch standard greenhouse pots, and then placed in a dark growth cabinet (36 total pots). After 7 d, the pots were placed under approximately 50 μmol m–2 s–1 light at room temperature (time 0). At varying times after illumination (2, 4, 12, and 48 h), the two newest leaves were collected from plants in two or three of the pots; these were randomly selected from the 36 pots. At each time point, plastids were isolated using a modification of established protocols (Leech and Leese, 1982). In brief, the leaf tissue was cut into small pieces, homogenized in a blender for 3 and then 5 s in 4 mL of isolation medium (0.067 m KH2PO4 [pH 8.0], 0.5 m Suc, 1 mm MgCl2, and 0.2% [w/v] bovine serum albumin) per gram of leaf tissue, and filtered through two layers of Miracloth (Calbiochem-Novabiochem, San Diego). The filtrate was then centrifuged for 90 s at 3,000g, and the pellets were decanted and frozen at –80°C. For each time point, 0.3 g fresh weight harvested tissue was saved in 80% (v/v) acetone for chlorophyll determinations by previously described methods (Aluru et al., 2001).
Isolation of Plastid Proteins
Plastid pellets were suspended in 20 mL of resuspension buffer (20 mm MOPS, 50 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride [pH 7.0]), and proteins were precipitated using 10% (v/v) trichloroacetic acid then washed twice with 100% (v/v) cold acetone. Samples were air dried overnight and dissolved the next day in rehydration buffer (7 m urea, 2 m thiourea, 4% [w/v] CHAPS, 40 mm Tris-Cl, 2 mm tributylphosphine (TBP), and 0.5% [w/v] carrier ampholytes added just before use). The protein samples were then stored at –80°C. Protein concentrations were determined using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories).
Two-Dimensional Gel Electrophoresis
IEF was performed using an IPGphor IEF System (Amersham-Pharmacia Biotech, Uppsala). Protein (125 μg) was mixed with rehydration buffer (final volume of 250 μL), and the samples were loaded onto 13-cm strips (pH 4–7) and rehydrated for 2 h at 20°C and 20 V for 10 h, 100 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 2,500 V for 1 h, and finally 8,000 V until the total V hours reached at least 80,000. After IEF, the strips were stored at –80°C. Before second dimension electrophoresis, the IEF strips were equilibrated in SDS equilibration buffer (50 mm Tris-Cl [pH 8.0], 6 m urea, 3% [w/v] SDS, 20% [v/v] glycerol, and 0.125% [v/v] concentrated tributylphosphine) for 30 min with gentle shaking. After equilibration, strips were applied to 12.5% (w/v) SDS-PAGE gels and sealed with agarose sealing solution (0.5% [w/v] agarose in SDS buffer plus a few grains of Bromphenol Blue). Protein samples were separated by SDS gel electrophoresis with running buffer (25 mm Tris, 192 mm Gly, and 0.1% [w/v] SDS). Protein Benchmark (Invitrogen, Carlsbad, CA) was applied to Whatman paper (Whatman, Clifton, NJ) and loaded as a molecular mass marker. Electrophoresis was carried out at 20 mA per gel with a maximum of 250 V for approximately 6 h. After electrophoresis, the gels were immediately stained with colloidal Coomassie Blue with gentle shaking for 2 d, then transferred to 1% (v/v) acetic acid destain with gentle shaking for 1 d. Next, the gels were transferred to new colloidal Coomassie stain for 1 d and then destain for 1 d. Finally, the gels were imaged using the PDQuest software on a GS-800 Calibrated Densitometer (Bio-Rad Laboratories). After imaging, the gels were stored in destain at 4°C. Spot intensities were determined using the software PDQuest.
Mass Spectrometry
Each spot was manually excised from the gel and placed into a microcentrifuge tube containing 50% (v/v) methanol. Each gel piece was then destained by washing two to three times with wash buffer (2.5 mm Tris-HCl [pH 8.5] and 50% [v/v] acetonitrile) and dried in a speed vacuum. Sequencing grade modified trypsin (5 μL; Promega, Madison, WI) was added to the dried gel slice and in gel digestion took place overnight while shaking at 37°C. Peptides were eluted from the gel piece using 5 μL of peptide elution buffer (50% [v/v] acetonitrile and 0.5% [v/v] trifluoroacetic acid). After centrifugation at 14,000 rpm for approximately 90 s, 1 μL of the eluted peptide mixture was mixed with the MALDI-TOF matrix (α-cyano-4-hydroxycinnamic acid in 50% [w/v] acetonitrile and 0.5% [w/v] trifluoroacetic acid), spotted onto a MALDI plate, and air dried. A Voyager-DE Pro MALDI-TOF mass spectrometer (Perspective Biosystems, Hertford, Great Britain) was used for mass spectrophotometric analysis.
After spectra were obtained, they were calibrated using Data Explorer software, version 4.0 (PE-Applied Biosystems, Foster City, CA). Internal standards, Angiotensen I (mass-to-charge ratio = 904.4681) and Bradykinin 2–9 (mass-to-charge ratio = 1296.6853), were included in the matrix solution, and the peaks were calibrated using these standards. For identification, the resulting peptide fingerprint was searched against bioinformatic databases using the software Ms-Fit version 3.3.1 from the software suite Protein Prospector version 3.4.1. The databases included NCBI nonredundant proteins limited to plants (http://www.ncbi.nlm.nih.gov) and TIGR assembled expressed sequence tags for maize (http://www.tigr.org). We developed software to search the databases in “batch” mode (see “Results”).
Once an identification was obtained, the spot was verified by matching the calculated molecular mass and pI against the actual experimental spot mass and pI. Spots were also verified by comparing the most intense peaks on the mass spectrum to the peptide mass fragments relied upon for identification. Although we found it useful to compare our gels with a proteome map of maize whole leaf tissue (Porubleva et al., 2001), this map and ours have a high spot density and were generated under different electrophoresis conditions. Hence, the two maps cannot be superimposed for exact protein spot identification purposes.
Data Analysis
PDQuest software was used to assemble first and second level match sets. A first level match set (standard gel) represents a “standard image” of four replicate two-dimensional gels for each time point. Each spot included on the standard gel met several criteria: It was present in at least three of the four gels, it was qualitatively consistent in size and shape in the replicate gels, and its quantity was within the linear range of the densitometer. In addition to “quantity” scores (based on spot density and area), the PDQuest software assigns “quality” scores to each gel spot. The quality scores provide a measure of how well the software is able to assess a quantity for a given spot and ranges from 0 to 100, based on five attributes: (a) good fit to the Gaussian distribution model, (b) streaking in the X direction, (c) streaking in the Y direction, (d) overlap of the spot with other spots, and (e) whether the peak intensity value of the spot is within the linear range of the scanner (Bio-Rad, 2000). We defined “low-quality” spots as those with a quality score less than 30; these spots were eliminated from further analysis. The remaining high-quality quantities were used to calculate the median value for a given spot, and this value was used as the spot quantity on the standard gel. After obtaining first level match sets, PDQuest was used to assemble a second level match set that allowed a comparison of the standard gels from each of the time points. From this match set, the filtered spot quantities from the standard gels were assembled into a data matrix of high-quality spots from the five time points.
Four statistical techniques were used to analyze the data. PAL cluster analysis and PCA were performed using the software TreeView version 1.5 and Cluster version 2.1.1, respectively (http://rana.lbl.gov/EisenSoftware.htm). We used a covariance matrix for the PCA analysis. We wrote software to perform ART2 clustering on normalized medians (see “Results”). SOM was performed on normalized medians using version 1.0 of Gene Cluster (http://www-genome.wi.mit.edu/cancer/software/software.html).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We would like to thank Xiaowu Gai (Laurence H. Baker Center for Bioinformatics and Biological Statistics, Iowa State University, Ames) for writing the batch program and Ericka Havecker (Iowa State University, Ames), Lawrence Bogorad (Harvard University, Cambridge, MA), and four anonymous reviewers for careful review of this manuscript.
This work was supported by the National Science Foundation (Integrative Graduate Education and Research Traineeship [IGERT] training grant in Bioinformatics and Computational Biology to P.L.) and by the U.S. Department of Energy (Energy Biosciences; grant no. DE–FG02–94ER20147 to S.R.).
References
- Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K et al. (2001) Chloroplast and mitochondrial proteases in Arabidopsis thaliana: a proposed nomenclature. Plant Physiol 125: 1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR (2001) The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol 127: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardel J, Louwagie M, Jaquinod M, Jourdain A, Luche S, Rabilloud T, Macherel D, Garin J, Bourguignon J (2002) A survey of the plant mitochondrial proteome in relation to development. Proteomics 2: 880–898 [DOI] [PubMed] [Google Scholar]
- Battey NH, Dickinson HG, Hetherington AM (1993) Some roles of posttranslational modifications in plants. In NH Battey, HG Dickinson, AM Hetherington, eds, Post-Translational Modifications in Plants. Cambridge University Press, Cambridge, UK, pp 1–16
- Bauer J, Hiltbrunner A, Kessler F (2001) Molecular biology of chloroplast biogenesis: gene expression, protein import and intraorganellar sorting. Cell Mol Life Sci 58: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C (1997) Cinnamate-4-hydroxylase expression in Arabidopsis: regulation in response to development and the environment. Plant Physiol 113: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bio-Rad Laboratories (2000) PDQuest User Guide for Version 6.2. Bio-Rad Laboratories, Hercules, CA
- Bogorad L (1991) Possibilities for intergenomic integration: regulatory crosscurrents between the plastid and nuclear-cytoplasmic compartments. Cell Cult Somatic Cell Genet Plants 7B: 447–466 [Google Scholar]
- Bringloe DH, Rao SK, Dyer TA, Raines CA, Bradbeer JW (1996) Differential gene expression of chloroplast and cytosolic phosphoglycerate kinase in tobacco. Plant Mol Biol 30: 637–640 [DOI] [PubMed] [Google Scholar]
- Carpenter GA, Grossber S, Rosen DB (1991) Art 2-A: an adaptive resonance algorithm for rapid category learning and recognition. Neural Networks 4: 493–504 [Google Scholar]
- Chen S, McMahon D, Bogorad L (1967) Early effects of illumination on the activity of some photosynthetic enzymes. Plant Physiol 42: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, Budworth PR (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR (2002) Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 23: 1754–1765 [DOI] [PubMed] [Google Scholar]
- Christie JM, Briggs WR (2001) Blue light sensing in higher plants. J Biol Chem 276: 11457–11460 [DOI] [PubMed] [Google Scholar]
- Cook WB, Walker JC (1992) Identification of a maize nucleic acid-binding protein (NBP) belonging to a family of nuclear-encoded chloroplast proteins. Nucleic Acids Res 20: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker G, Wanner G, Zenk MH, Lottspeich F (2000) Characterization of proteins in latex of the opium poppy (Papaver somniferum) using two-dimensional gel electrophoresis and microsequencing. Electrophoresis 21: 3500–3516 [DOI] [PubMed] [Google Scholar]
- de Heij HT, Jochemsen AG, Willemsen PT, Groot GS (1984) Protein synthesis during chloroplast development in Spirodela oligorhiza: coordinated synthesis of chloroplast-encoded and nuclear-encoded subunits of ATPase and ribulose-1,5-bisphosphate carboxylase. Eur J Biochem 138: 161–168 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Conley TR, Shih MC, Goodman HM (1993) Effects of blue and red light on expression of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase of Arabidopsis thaliana. Plant Physiol 103 1115–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo J, Zanders E, Dragoni I, Amphlett G, Falciani F (2001) Methods and approaches in the analysis of gene expression data. J Immunol Methods 250: 93–112 [DOI] [PubMed] [Google Scholar]
- Drzymalla C, Schroda M, Beck CF (1996) Light-inducible gene HSP70B encodes a chloroplast-localized heat shock protein in Chlamydomonas reinhardtii. Plant Mol Biol 31: 1185–1194 [DOI] [PubMed] [Google Scholar]
- Esen A (1992) Purification and partial characterization of maize (Zea mays L.) beta-glucosidase. Plant Physiol 98: 74–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Forger JM, Bogorad L (1973) Steps in the acquisition of photosynthetic competence by plastids of maize. Plant Physiol 52: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant SI (1993) Neural Network Learning and Expert Systems. MIT Press, Cambridge, MA
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M (1998) Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol 177: 115–180 [DOI] [PubMed] [Google Scholar]
- Grebanier AE, Steinback KE, Bogorad L (1979) Comparison of the molecular weights of proteins synthesized by isolated chloroplasts with those which appear during greening in Zea mays. Plant Physiol 63: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Aebersold R (2001) Advances in proteome analysis by mass spectrometry. J Biol Chem 276: 45497–45500 [DOI] [PubMed] [Google Scholar]
- Groth G, Strotmann H (1999) New results about structure, function and regulation of the chloroplast ATP synthase (CF0CF1). Physiol Plant 106: 142–148 [Google Scholar]
- Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W (2000) The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21: 1037–1053 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold SA (1999) Quantitative analysis of protein mixtures using isotope coded affinity tags. Nat Biotechnol 17: 994–999 [DOI] [PubMed] [Google Scholar]
- Hadjiiski L, Sahiner B, Chan HP, Petrick N, Helvie M (1999) Classification of malignant and benign masses based on hybrid ART2LDA approach. IEEE Trans Med Imaging 18: 1178–1187 [DOI] [PubMed] [Google Scholar]
- Hankamer B, Barber J, Boekema EJ (1997) Structure and membrane organization of photosystem II in green plants. Annu Rev Plant Physiol Plant Mol Biol 48: 641–671 [DOI] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ (1988) Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333: 330–334 [DOI] [PubMed] [Google Scholar]
- Herbert B, Righetti PG (2000) A turning point in proteome analysis: sample prefractionation via multicompartment electrolyzers with isoelectric membranes. Electrophoresis 21: 3639–3648 [DOI] [PubMed] [Google Scholar]
- Hippler M, Klein J, Fink A, Allinger T, Hoerth P (2001) Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J 28: 595–606 [DOI] [PubMed] [Google Scholar]
- Hubbard MJ (2002) Functional proteomics: the goalposts are moving. Proteomics 2: 1069–1078 [DOI] [PubMed] [Google Scholar]
- Jolliffe IT (1986) Principal Component Analysis. Springer-Verlag, New York
- Ke J, Wen T-N, Nikolau BJ, Wurtele ES (2000) Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heterotrimeric acetyl-coenzyme A carboxylase. Plant Physiol 122: 1057–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach T, Bystedt M, Hynds P, Robinson C, Schroder WP (2000) A peroxidase homologue and novel plastocyanin located by proteomics to the Arabidopsis chloroplast thylakoid lumen. FEBS Lett 480: 271–276 [DOI] [PubMed] [Google Scholar]
- Komatsu S, Muhammad A, Rakwal R (1999) Separation and characterization of proteins from green and etiolated shoots of rice (Oryza sativa L.): towards a rice proteome. Electrophoresis 20: 630–636 [DOI] [PubMed] [Google Scholar]
- Konishi H, Komatsu S (2003) A proteomics approach to investigating promotive effects of brassinolide on lamina inclination and root growth in rice seedlings. Biol Pharmacol Bull 26: 401–408 [DOI] [PubMed] [Google Scholar]
- Koumoto Y, Shimada T, Kondo M, Takao T, Shimonishi Y, Hara-Nishimura I, Nishimura M (2001) Chloroplast Cpn20 forms a tetrameric structure in Arabidopsis thaliana. Plant J 17: 467–477 [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun H-P (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol 127: 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Landick R, Oxender DL (1985) The complete nucleotide sequences of the Escherichia coli LIV-BP and LS-BP genes: implications for the mechanism of high-affinity branched-chain amino acid transport. J Biol Chem 260: 8257–8261 [PubMed] [Google Scholar]
- Leech RM, Leese BM (1982) Isolation of etioplasts from maize. In M Edelman, RB Hallick, eds, Methods in Chloroplast Molecular Biology. Elsevier Biomedical Press, New York, pp 221–233
- Leister D (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Leon P, Arroyo A, Mackenzie S (1998) Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Physiol Plant Mol Biol 49: 453–480 [DOI] [PubMed] [Google Scholar]
- Li J, Brick P, O'Hare MC, Skarzynski T, Lloyd LF, Curry VA, Clark IM, Bigg HF, Hazleman BL, Cawston TE et al. (1995) Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller. Structure 3: 541–549 [DOI] [PubMed] [Google Scholar]
- Liveanu V, Yocum CF, Nelson N (1986) Polypeptides of the oxygen-evolving photosystem II complex: immunological detection and biogenesis. J Biol Chem 261: 5296–5300 [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Mann M, Hendrickson RC, Pandey A (2001) Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem 70: 437–473 [DOI] [PubMed] [Google Scholar]
- Martel R, Cloney LP, Pelcher LE, Hemmingsen SM (1990) Unique composition of plastid chaperonin-60: α and β polypeptide-encoding genes are highly divergent. Gene 94: 181–187 [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG (1998) Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol 118: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver C (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Molloy MP (2000) Two-dimensional electrophoresis of membrane proteins using immobilized pH gradients. Anal Biochem 280: 1–10 [DOI] [PubMed] [Google Scholar]
- Ndimba BK, Chivasa S, Hamilton JM, Simon WJ, Slabas AR (2003) Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegsatra K (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O, Adam Z (1996) Effects of light and temperature on expression of ClpC, the regulatory subunit of chloroplastic Clp protease, in pea seedlings. Plant Mol Biol 31: 673–676 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12: 319–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubleva L, Vander Velden K, Kothari S, Oliver DJ, Chitnis PR (2001) The proteome of maize leaves: use of gene sequences and expressed sequence tag data for identification of proteins with peptide mass fingerprints. Electrophoresis 22: 1724–1738 [DOI] [PubMed] [Google Scholar]
- Prime T, Sherrier DJ, Mahon P, Packman LC, Dupree P (2000) A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499 [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Blisnick T, Heller M, Luche S, Aebersold R, Lunardi J, Braum-Breton C (1999) Analysis of membrane proteins by two-dimensional electrophoresis: comparison of the proteins extracted from normal or Plasmodium falciparum-infected erythrocyte ghosts. Electrophoresis 20: 3603–3610 [DOI] [PubMed] [Google Scholar]
- Rakwal R, Komatsu S (2000) Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis 21: 2492–2500 [DOI] [PubMed] [Google Scholar]
- Robertson D, Mitchell GP, Gilroy JS, Gerrish C, Bolwell GP, Slabas AR (1997) Differential extraction and protein sequencing reveals major differences in patterns of primary cell wall proteins from higher plants. J Biol Chem 272: 15841–15848 [DOI] [PubMed] [Google Scholar]
- Rodermel S (2001) Pathways of plastid-to-nucleus signaling. Trends Plant Sci 6: 471–478 [DOI] [PubMed] [Google Scholar]
- Rodermel SR, Bogorad L (1985) Maize plastid photogenes: mapping and photoregulation of transcript levels during light-induced development. J Cell Biol 100: 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel SR, Bogorad L (1987) Molecular evolution and nucleotide sequences of the maize plastid genes for the alpha subunit of CF1 (atpA) and the proteolipid subunit of CF0 (atpH). Genetics 116: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB (1994) Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene. Plant Physiol 105: 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni V, Kieffer S, Desclaux D, Masson F, Rabilloud T (2000) Membrane proteomics: use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis 21: 3329–3344 [DOI] [PubMed] [Google Scholar]
- Santoni V, Rouquie D, Doumas P, Mansion M, Boutry M, Degand H, Dupree P, Packman L, Sherrier J, Prime Y et al. (1998) Use of a proteome strategy for tagging proteins present at the plasma membrane. Plant J 16: 633–641 [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J (2000) Travelling of proteins through membranes. Planta 211: 449–456 [DOI] [PubMed] [Google Scholar]
- Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T (2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem 277: 8354–8365 [DOI] [PubMed] [Google Scholar]
- Sheen J-Y, Bogorad L (1985) Differential expression of the ribulose bisphosphate carboxylase large subunit gene in bundle sheath and mesophyll cells of developing maize leaves is influenced by light. Plant Physiol 79: 1072–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Jing Y, Kuang T (2003) Proteomics approach to identify wound-response related proteins from rice leaf sheath. Proteomics 3: 527–535 [DOI] [PubMed] [Google Scholar]
- Sherlock G (2000) Analysis of large-scale gene expression data. Curr Opin Immunol 12: 201–205 [DOI] [PubMed] [Google Scholar]
- Skylas DJ, Copeland L, Rathmell WG, Wrigley CW (2001) The wheat-grain proteome as a basis for more efficient cultivar identification. Proteomics 1: 1542–1546 [DOI] [PubMed] [Google Scholar]
- Smith AD, Sinha A (1999) Unsupervised classification of Space Acceleration Measurement System (SAMS) data using ART2-A. Microgravity Sci Technol 12: 91–100 [PubMed] [Google Scholar]
- Strzalka K, Tsugeki R, Nishimura M (1994) Heat shock induces synthesis of plastid-associated hsp70 in etiolated and greening pumpkin seedlings. Folia Histochem Cytobiol 32: 45–49 [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL (2001) Comprehensive expression profile of the Arabidopsis Hsp70 gene family. Plant Physiol 126: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell 14: S327–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ (2000) Proteomics of the chloroplast: experimentation and prediction. Trends Plant Sci 5: 420–425 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ (2001) Challenges and prospects of plant proteomics. Plant Physiol 126: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Asirvatham VS, Wang L, Sumner LW (2003) Mapping the proteome of barrel medic (Medicago trunculata). Plant Physiol 131: 1104–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KA, McManus MT, Gordon ME, Jordan TW (2002) The proteomics of senescence in leaves of white clover, Trifolium repens (L). Proteomics 2: 1114–1122 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Prieto S, Beligni M, Haynes PA, McDonald WH, Yates JR, Mayfield S (2002) Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14: 2957–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins: identification of all the proteins in the 50S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR (2000) The plastid ribosomal proteins: identification of all the proteins in the 30S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28455–28465 [DOI] [PubMed] [Google Scholar]