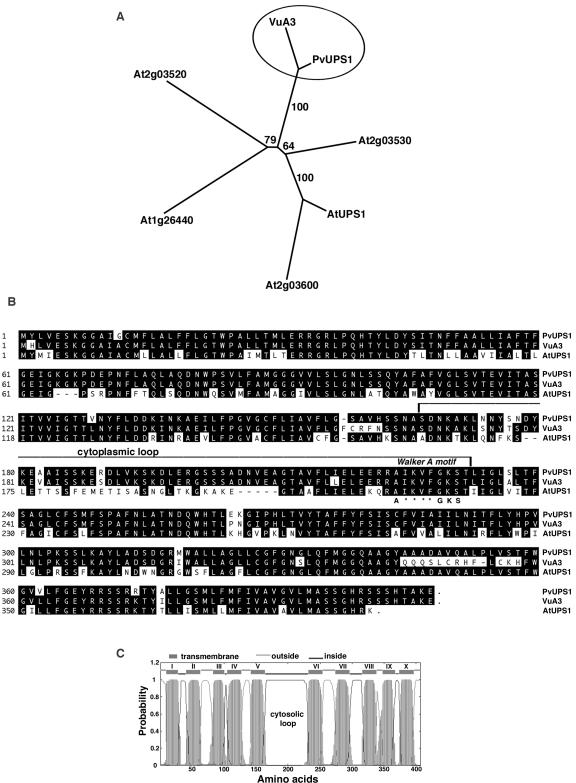
Figure 1.
Analysis of PvUPS1 and related proteins. A, Phylogenetic analysis. The tree is based on maximum parsimony analysis (PAUP 4.0b5, Rogers and Swofford, 1998) of aligned protein sequences from French bean (PvUPS1), cowpea (VuA3 protein/X90487), and Arabidopsis (AtUPS1/At2g03590, At2g03530, At2g03600, At2g03520, and At1g26440). The numbers indicate the occurrence of a given branch in 10,000 bootstrap replicates of the given data set. B, Amino acid sequence of PvUPS1 and alignment with two related members of the UPS family. Using PvUPS1 from French bean, AtUPS1 from Arabidopsis (At2g03590) and VuA3 from cowpea (X90487), a sequence alignment was generated by Clustal algorithm (LASERGENE software; DNASTAR, Madison, WI). Black background shows identical amino acids. Dashes indicate gaps in the sequences to allow maximal alignment. The third cytoplasmic loop is highlighted as well as the “Walker A” motif (A-x(4)-G-K-S), in which asterisks refer to variable amino acids. C, Topology of PvUPS1 using a transmembrane prediction program (TMHMM version 2.0; http://www.cbs.dtu.dk/services). PvUPS1 contains a large central loop probably located in the cytosol. The C and N terminus are predicted to be outside.