Figure 1. Methylation patterns of COBRALINE-1.
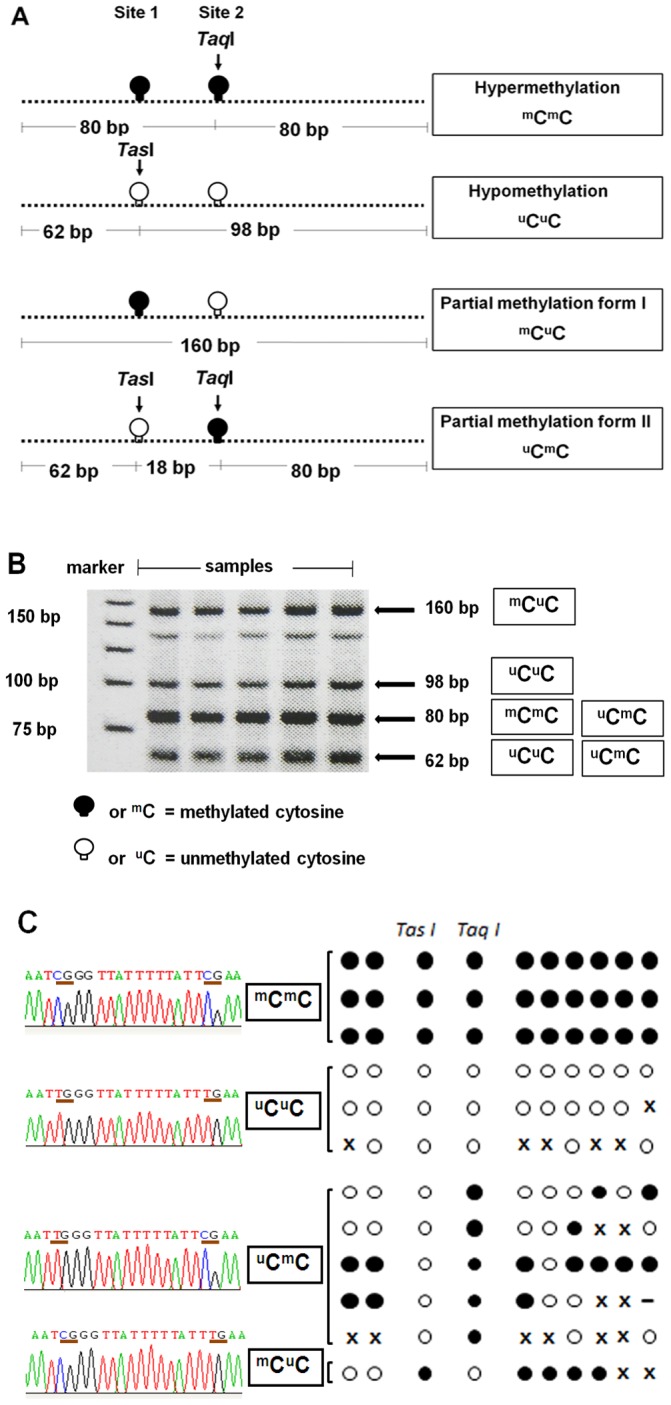
(A) The LINE-1 amplicons were 160 bp and had 2 CpG dinucleotides. Four patterns of methylated CpGs were detected, including hypermethylation (mCmC), hypomethylation (uCuC), and two forms of partial methylation (mCuC and uCmC). The TasI enzyme targets unmethylated cytosine site 1, and TaqI targets methylated cytosine site 2. (B) After restriction digestion with TasI and TaqI, four sizes of products (160, 98, 80, and 62 bp) were identified, depending on the methylation status of both CpG loci. (C) Examples of bisulfite sequencing. Left side represents sequences of the two CpG dinucleotides at TasI and TaqI cut site whereas the right side represents the CpG dinucleotides in the amplified PCR products. Each circle exemplifies the methylation status of each selected clone. Black and white circles are methylated and unmethylated CpG dinucleotides, respectively. x is mutated sequence and – is deleted sequence.
