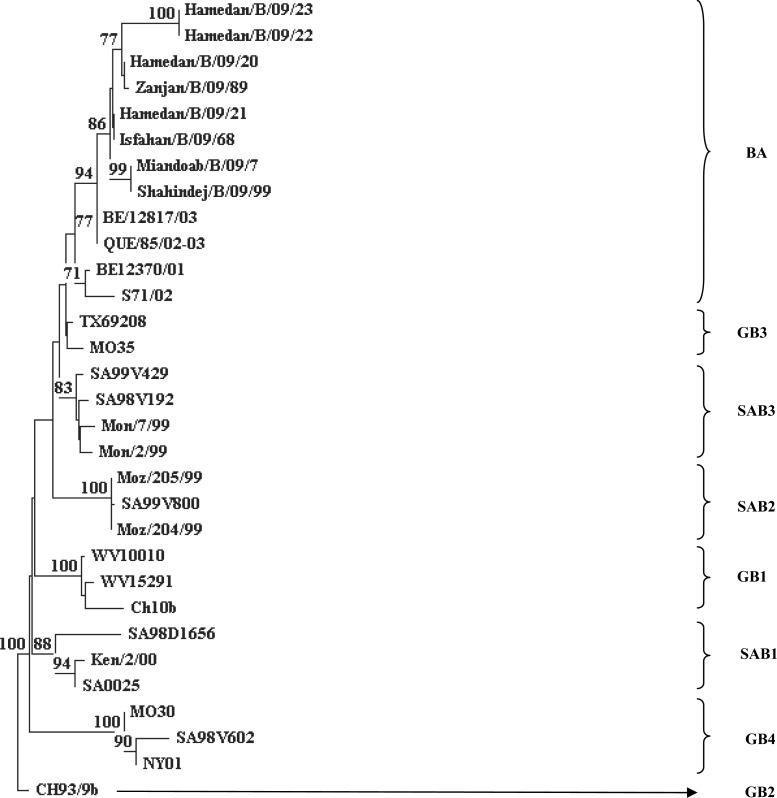
Fig. 2.
Phylogenetic tree constructed with the HRSV subgroup B nucleotide sequences of based on the second variable region of the G gene. The nucleotide sequences were aligned with the CLUSTAL X (version 1/83), and phylogenetic tree were prepared with TREECON software with a neighbor-joining algorithm. Pairwise distances between and within the genotypes were calculated with Kimura 2 parameters. Iranian isolates indicated by city/subtype/year/sample number. The following reference sequences were used to construct tree: WV10010, WV15291, and CH10b (GB1); CH93-9b (GB2); MO35, and TX69208(GB3);MO30, NY01, and SA98V602 (GB4); SA98D1656, SA0025, and Ken/2/00 (SAB1); SA99V800, Moz/204/99, and Moz/205/99 (SAB2); SA99V429, SA98V192, Mon/7/99, and Mon/2/99 (SAB3): S71/02, QUE/85/02-03, BE/12370/01, and BE/12817/03 (BA).