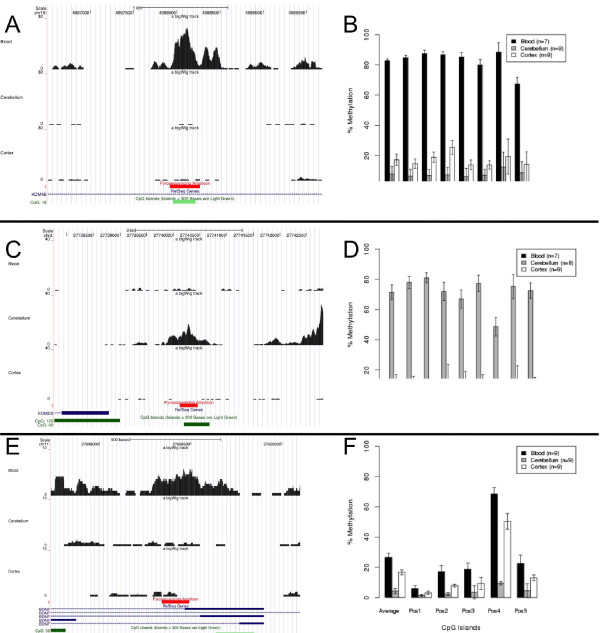
Figure 3.
Verification and replication of MeDIP-seq data for three top-ranked CGI DMRs. (a, b) Tissue-specific DNA methylation across an intragenic CGI in the JMJD2B/KDM4B gene. (a) MeDIP-seq analysis shows this region is hypermethylated in blood DNA compared to cortex and cerebellum (the red bar depicts the region subsequently analyzed by bisulfite pyrosequencing). (b) Pyrosequencing data for this region in an extended sample set confirm significant tissue-specific methylation patterns (P = 2 × 10-8). (c, d) Tissue-specific DNA methylation across a CGI in the promoter of the EOMES gene. (c) MeDIP-seq analysis shows this region is hypermethylated in cerebellum DNA compared to cortex and blood. (d) Pyrosequencing data for this region in an extended sample set confirm significant tissue-specific methylation patterns (P = 2 × 10-5). (e, f) Tissue-specific DNA methylation across an intragenic CGI in the BDNF gene. (e) MeDIP-seq analysis shows this region is hypermethylated in blood DNA compared to cortex and cerebellum from the same individuals. (f) Pyrosequencing data for this region in an extended sample set confirm significant tissue-specific methylation patterns (P = 4 × 10-9). Error bars represent standard error of the mean.