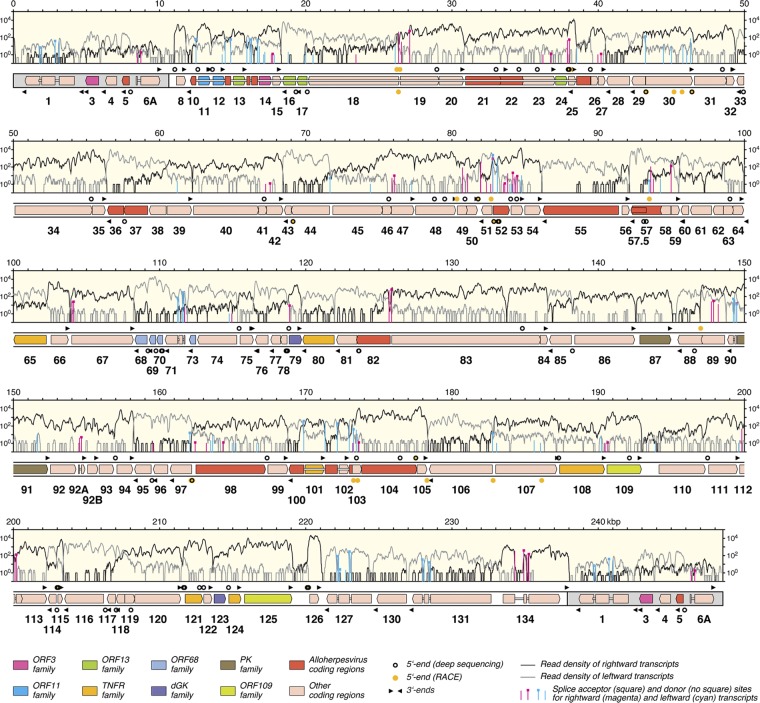
Fig 1.
Map of the AngHV1 genome showing transcription and splicing of ORFs predicted to encode functional proteins. The terminal direct repeats are shaded gray. ORFs are depicted as color-shaded arrows, with names (lacking the ORF prefix) below. The ORF colors indicate conservation among alloherpesviruses or families of related genes (see the key). Introns connecting spliced ORFs are shown as narrow white bars. The locations of transcript 5′ and 3′ ends (Tables 2 and 3) are marked, rightward above the genome and leftward below. The light-yellow windows contain the transcriptome profile as two traces, separated into rightward (black) and leftward (gray) transcripts. The vertical lines indicate the locations of splice donor and acceptor sites supported by >10 reads (Table 1; also see data set S1 [set II] in the supplemental material), divided into rightward (magenta) and leftward (cyan) splicing. The height of each line indicates the number of reads supporting transcription of the splice site, plotted on a log10 scale.