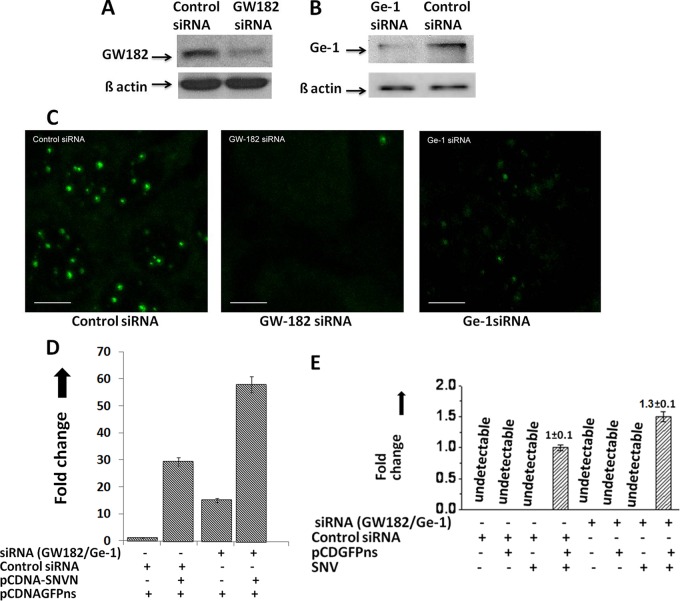
Fig 5.
Role of cellular P bodies in hantavirus cap snatching. (A) Downregulation of GW182 by siRNA. To determine whether GW182 was downregulated after siRNA treatment, cells transfected with either siRNA or control siRNA were lysed and GW182 expression was monitored by Western blot analysis using anti-GW182 antibody. To confirm that downregulation of GW182 observed by Western blot analysis was not a loading error, equal volumes of cell lysate were examined for the expression of β-actin by Western blot analysis (bottom bands). (B) Similar to what is shown in panel A, the downregulation of Ge-1 by siRNA was confirmed by Western blot analysis using anti-Ge-1 antibody. (C) Downregulation of GW182 and Ge-1 by siRNA causes the downregulation of cellular P bodies. Huh-7 cells were transfected with 150 nM either control siRNA, GW-182 siRNA, or Ge-1 siRNA. Twenty-four hours posttransfection, cells were fixed with formaldehyde and stained with anti-Dcp2 antibody. Cells were visualized under fluorescence microscope using a FITC-conjugated secondary antibody. Bar, 10 μm. (D) Wild-type or P-body-downregulated Huh-7 cells were cotransfected with pCDNAGFPns and pCDNA-SNVN plasmids, expressing the cap donor test mRNA and SNV N protein, respectively. Thirty-six hours posttransfection, cells were lysed and total RNA was purified and reverse transcribed using random primers. The effect of N upon the stability of the test mRNA was quantified by real-time PCR analysis using a forward primer, 5′-TAGAGAACCCACTGCTTACTGGC-3′, and a reverse primer, 5′-CAGATGAACTTCAGGGTCAG-3′, to amplify 241 nucleotides from the 5′ mRNA terminus. (E) Huh-7 cells transfected with either siRNA or control siRNA along the cap donor plasmid pCDNAGFPns were infected with SNV. Caps snatched by the SNV from the test mRNA were quantified by real-time PCR analysis using the primer set shown in Fig. 1A.